For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSGLRELCRCGRGLLRHRRSGTRGGPQDGGSFRGQDPGRGGCAQLSGGRVGSRRRMTSRVVIVGSSVGGV RTAQALRSQGYDGDIVLVGEETALPYDKPPLSKALLAGTSDIAAATLLTRDAAAADAIELLLGRRAIGVD VAACQVEFADDEPLRYDHLVVATGASARPSPWGHGSGVHVLRTLEDCLRLRADLVAGARLVVIGAGFIGA EVASTARSLGLQVTVVDPVPVPMSRVLTEEIGGWFVDLHRDHGVHTRFGVGVENIEGERGNLKIGLTDGT VLEADTVVVGIGAVPNDGWLMPSGLVVDDGLVCDEFCRAVTSQNIYAVGDVSRWFHRARGVTMRIEHWTN AVDQASCVAHNILYPDELRCYEPVEYVWSDQYDWKIQFCGQTTHLGEPLVVGDRLSDNRFTALYPDREQR LSGAAIVNWPRALIECRRALKAGSDLETVRERLELLTNPPDTAATRSP
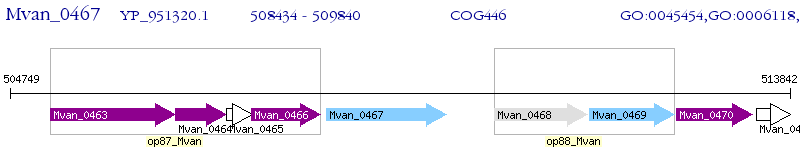
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0467 | - | - | 100% (468) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1290 | - | 7e-69 | 40.05% (392) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1327 | - | 2e-58 | 37.89% (388) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0707 | - | 4e-66 | 39.14% (396) | putative ferredoxin reductase |
| M. gilvum PYR-GCK | Mflv_0680 | - | 0.0 | 97.58% (413) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv0688 | - | 4e-66 | 39.14% (396) | putative ferredoxin reductase |
| M. leprae Br4923 | MLBr_02387 | lpd | 5e-09 | 26.34% (205) | dihydrolipoamide dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3838c | - | 2e-67 | 39.03% (392) | putative ferredoxin reductase |
| M. marinum M | MMAR_1017 | - | 5e-71 | 41.67% (384) | ferredoxin reductase |
| M. avium 104 | MAV_4485 | - | 4e-66 | 40.55% (397) | ferredoxin reductase |
| M. smegmatis MC2 155 | MSMEG_1416 | - | 5e-70 | 40.92% (391) | pyridine nucleotide-disulphide oxidoreductase |
| M. thermoresistible (build 8) | TH_3739 | - | 2e-71 | 40.15% (396) | PUTATIVE Pyridine nucleotide-disulphide oxidoreductase |
| M. ulcerans Agy99 | MUL_0769 | - | 4e-71 | 41.67% (384) | ferredoxin reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0467|M.vanbaalenii_PYR-1 MSGLRELCRCGRGLLRHRRSGTRGGPQDGGSFRGQDPGRGGCAQLSGGRV
Mflv_0680|M.gilvum_PYR-GCK --------------------------------------------------
Mb0707|M.bovis_AF2122/97 -------------------------------------MNAHVTSREGVN-
Rv0688|M.tuberculosis_H37Rv -------------------------------------MNAHVTSREGVN-
MMAR_1017|M.marinum_M ------------------------------------MYEHHVTSSEGVTQ
MUL_0769|M.ulcerans_Agy99 ------------------------------------MYEHHVTSSEGVTQ
MAV_4485|M.avium_104 -------------------------------------MSSDASAS-----
MSMEG_1416|M.smegmatis_MC2_155 -------------------------------------MTTSAHSA-----
TH_3739|M.thermoresistible__bu -------------------------------------VSTT---------
MAB_3838c|M.abscessus_ATCC_199 -------------------------------------MTDAER-------
MLBr_02387|M.leprae_Br4923 -------------------------------MTHYDVVVLGAGPGGYVAA
Mvan_0467|M.vanbaalenii_PYR-1 GSRRRMTSRVVIVG----SSVGGVRTAQALRSQGYDGDIVLVGEETALPY
Mflv_0680|M.gilvum_PYR-GCK -----MTSRVVIVG----SSVGGVRTAQALRSQGYDGGIVLVGEETALPY
Mb0707|M.bovis_AF2122/97 ----EFDDGIVIVG----GGLAAARTAEQLRRAGYSGRLTIVSDEVHLPY
Rv0688|M.tuberculosis_H37Rv ----EFDDGIVIVG----GGLAAARTAEQLRRAGYSGRLTIVSDEVHLPY
MMAR_1017|M.marinum_M DPGATAANGIVIIG----GGLAAARTAEQLRRAGYEGQLTIVSDEVHLPY
MUL_0769|M.ulcerans_Agy99 DPGATAANGIVIIG----GGLAAARTAEQLRRAGYEGQLTIVSDEVHLPY
MAV_4485|M.avium_104 -------NGIVIVG----GGLAAARTAEQLRRSEYSGPITIVSDEVHLPY
MSMEG_1416|M.smegmatis_MC2_155 --------GIVIVG----GGLAATRTAEQLRRSEYAGPITIVSDEDHLPY
TH_3739|M.thermoresistible__bu -------AGIIIVG----GGLAAARTAEQLRRSEFGGPITIVSDEDHLPY
MAB_3838c|M.abscessus_ATCC_199 --------GVLIIG----GGLGAVRTAEQLRRAEFTGPITIVSSETHLPY
MLBr_02387|M.leprae_Br4923 IRAAQLGLSTAVVEPKYWGGICLNVGCIPSKVLLHNAELAHIFTKEAKTF
:: ..: . : . . :. : : .:
Mvan_0467|M.vanbaalenii_PYR-1 DKP---PLSKALLAG-TSDIAAATLLTRDAAAADAIELLLGRRAIGVDVA
Mflv_0680|M.gilvum_PYR-GCK DKP---PLSKALLAG-TSDIAAATLLTRDAAAADAIELLLGRRAIGVDVA
Mb0707|M.bovis_AF2122/97 DRP---PLSKEVLR---SEVDDVALKPREFYDEKDIALRLGSAAVSLDTG
Rv0688|M.tuberculosis_H37Rv DRP---PLSKEVLR---SEVDDVALKPREFYDEKDIALRLGSAAVSLDTG
MMAR_1017|M.marinum_M DRP---PLSKEVLR---SEVDDVSLKPREWYDEKDIALRLGSAATGLDTA
MUL_0769|M.ulcerans_Agy99 DRP---PLSKEVLR---SEVDDVSLKPREWYDEKGITLRLGSAATGLDTA
MAV_4485|M.avium_104 DRP---PLSKEVLR---KEVDDTALKPRQWYDDNDITLRLGSAARSLDTA
MSMEG_1416|M.smegmatis_MC2_155 DRP---PLSKEVLR---AENDDVTLKPAEFYEENNITLRLGSGAQSVDTA
TH_3739|M.thermoresistible__bu DRP---PLSKDVLS---GKTDDVTLKPAEFYAENDITLLLGTAATALDPQ
MAB_3838c|M.abscessus_ATCC_199 DRP---PLSKDVLRDAEKTIDAVVLKPREFYDEKQIELRLGSEVTALDPV
MLBr_02387|M.leprae_Br4923 GISGDASFDYGIAYDRSRKVSEGRVAGVHFLMKKNKITEIHGYGRFTDAN
. . .:. : : . . : *
Mvan_0467|M.vanbaalenii_PYR-1 ACQVEFADDE-----PLRYDHLVVATGASARPSP-WGHGSGVHVLRTLED
Mflv_0680|M.gilvum_PYR-GCK ACQVEFADDE-----PLHYDHLVVATGASARPSP-WGHGSGVHVLRTLED
Mb0707|M.bovis_AF2122/97 EQTVTLADGT-----VLGYDELVIATGLVPRRIPSLPDLDGIRVLRSFDE
Rv0688|M.tuberculosis_H37Rv EQTVTLADGT-----VLGYDELVIATGLVPRRIPSLPDLDGIRVLRSFDE
MMAR_1017|M.marinum_M AQTVTLADGT-----VLGYDELVIATGLVPRRIPAFPELEGIRVLRSFDE
MUL_0769|M.ulcerans_Agy99 AQTVTLADGT-----VLGYDELVIATGLVPRRIPAFPELEGIRVLRSFDE
MAV_4485|M.avium_104 AQTLTLEDGT-----TLGYDELVIATGLVPRRIPTIPDLDGIRVLRTFDE
MSMEG_1416|M.smegmatis_MC2_155 AQTVRLADGS-----ELPYDELVIATGLVPKRIGSFPDLQGIRVLRTYDE
TH_3739|M.thermoresistible__bu ARKLTLADGR-----ALEYDELIIATGLVPKRIPTLPDLEGIRVLRTYDE
MAB_3838c|M.abscessus_ATCC_199 ARTVTLSDGG-----TLEYGEVIIATGLVPRRIPTFPDLAGVHVLRTADD
MLBr_02387|M.leprae_Br4923 TLSVELSEGVPETPLKVTFNNVIIATGSKTRLVP--GTLLSTNVITYEEQ
: : :. : :..:::*** .: . .*: ::
Mvan_0467|M.vanbaalenii_PYR-1 CLRLRADLVAGARLVVIGAGFIGAEVASTARSLGLQVTVVDPVPVPMSRV
Mflv_0680|M.gilvum_PYR-GCK CLRLRADLVAGARLVVIGAGFIGAEVASTARSLGLQVTVVDPVPVPMSRV
Mb0707|M.bovis_AF2122/97 SMALRKHASAARHAVVVGAGFIGCEVAASLRGLGVDVVLVEPQPAPLASV
Rv0688|M.tuberculosis_H37Rv SMALRKHASAARHAVVVGAGFIGCEVAASLRGLGVDVVLVEPQPAPLASV
MMAR_1017|M.marinum_M CMALRSHASAAKRAVVIGAGFIGCEVAASLRGLGVEVVLVEPQPTPLAAV
MUL_0769|M.ulcerans_Agy99 CMALRSHASAAKRAVVIGAGFIGCEVAASLRGLGVEVVLVEPQPTPLAAV
MAV_4485|M.avium_104 SLALREHASAAQRAVIIGAGFIGCEVAASLRSLGVDVVLVEPQPTPLAAV
MSMEG_1416|M.smegmatis_MC2_155 SMALREDARKASRAVVIGAGFIGCEVAASLRKLGVDVVLVEPQPTPLASV
TH_3739|M.thermoresistible__bu SMALRKHAGSARNAVVIGAGFIGCEVAASLRGLGVHVVLVEPQPTPLAGV
MAB_3838c|M.abscessus_ATCC_199 SFALRGDAENARRAVVVGAGFIGCEVAATLRANGVEVVLVEPQPAPLLAA
MLBr_02387|M.leprae_Br4923 ILTR----ELPDSIVIVGAGAIGIEFGYVLKNYGVDVTIVEFLPRAMPNE
: *::*** ** *.. : *:.*.:*: * .:
Mvan_0467|M.vanbaalenii_PYR-1 LTEEIGGWFVDLHRDHGVHTRFGVGVENIEGERGNLKIGLTDG---TVLE
Mflv_0680|M.gilvum_PYR-GCK LTEEIGGWFVDLHRDHGVHTRFGVGVENIEGERGNLKIGLTDG---TVLE
Mb0707|M.bovis_AF2122/97 LGEQIGQLVTRLHRDEGVDVRTGVTVAEVRGKGHVDAVVLTDG---TELP
Rv0688|M.tuberculosis_H37Rv LGEQIGQLVTRLHRDEGVDVRTGVTVAEVRGKGHVDAVVLTDG---TELP
MMAR_1017|M.marinum_M LGEQIGELVARLHRSEGVDVRTGVGVAEVRGDGHVDTVVLADG---TQLD
MUL_0769|M.ulcerans_Agy99 LGEQIGELVARLHRSEGVDVRTGVGVAEVRGDGHVDTVVLADG---TQLD
MAV_4485|M.avium_104 LGQQIGELVARLHRAEGVDVRLGVGVSEVRGDRHVEAVVLTDG---TELA
MSMEG_1416|M.smegmatis_MC2_155 LGEKIGALVTRLHRAEGVDVRCGVGVTEVRGTQNVETVVLADG---TELE
TH_3739|M.thermoresistible__bu LGEQIGALVARLHRDEGVDLRCGVGVADVRGTGAVEKVVLDDG---TELD
MAB_3838c|M.abscessus_ATCC_199 IGQQLGDLVARLHRNEGVDVRVGVGVDAVEGQDRVQSVTLSDG---TRLD
MLBr_02387|M.leprae_Br4923 D-AEVSKEIEKQFKKMGIKILTGTKVESISDNGSHVLVAVSKDGQFQELK
::. . .: *:. *. * : . : : .. *
Mvan_0467|M.vanbaalenii_PYR-1 ADTVVVGIGAVPNDGWLMPSGLVVD---DGLVCDEFCRAVTSQNIYAVGD
Mflv_0680|M.gilvum_PYR-GCK ADTVVVGIGAVPNDGWLMPSGLVVD---DGLVCDEFCRAVTSQNIYAVGD
Mb0707|M.bovis_AF2122/97 ADLVVVGIGSTPATEWLEGSGVEVD---NGVICDKAG-RTSAPNVWALGD
Rv0688|M.tuberculosis_H37Rv ADLVVVGIGSTPATEWLEGSGVEVD---NGVICDKAG-RTSAPNVWALGD
MMAR_1017|M.marinum_M ADLVVVGIGSHPATGWLDGSGIAVD---NGVVCDAAG-RTSAPNVWALGD
MUL_0769|M.ulcerans_Agy99 ADLVVVGIGSHPATGWLDGSGIAVD---NGVVCDAAG-RTSAPNVWAFGD
MAV_4485|M.avium_104 ADVVVIGIGSRPATDWLEGSGVAIDSVDRGVLCDEAG-RTSAPNVWALGD
MSMEG_1416|M.smegmatis_MC2_155 ADLVIVGIGSRPSVDWLKGSGIEVD---NGVVCDEVG-RTSAPHVWAIGD
TH_3739|M.thermoresistible__bu ADLVVVGIGSTPATDWLQGSGVEVD---NGVVCDAVG-RTNVEHVWAIGD
MAB_3838c|M.abscessus_ATCC_199 TDLVVLGIGSRPAIDWLDGSGVAVD---NGVVCDAVG-ATGTPHVWALGD
MLBr_02387|M.leprae_Br4923 ADKVLQAIGFAPNVDGYGLDKVGVALTADKAVDIDDYMQTNVSHIYAIGD
:* *: .** * . : : : . :::*.**
Mvan_0467|M.vanbaalenii_PYR-1 VSRWFHRARGVTMRIEHWTNAVDQASCVAH---------NILYPDELRCY
Mflv_0680|M.gilvum_PYR-GCK VSRWFHRARGVTMRIEHWTNAVDQASCVAH---------NILYPDELRCY
Mb0707|M.bovis_AF2122/97 VASWRDPM-GHQARVEHWSNVADQARVVVP---------AMLGTDVPT-G
Rv0688|M.tuberculosis_H37Rv VASWRDPM-GHQARVEHWSNVADQARVVVP---------AMLGTDVPT-G
MMAR_1017|M.marinum_M VASWRDQM-GHQVRVEHWSNVADQARVVVP---------AMLGREVSS-N
MUL_0769|M.ulcerans_Agy99 VASWRDQM-GHQVRVEHWSNVADQARVVVP---------AMLGREVSS-N
MAV_4485|M.avium_104 VASWRDAT-GHQGRVEHWSNVADQARVVVP---------AMLGKEVPP-V
MSMEG_1416|M.smegmatis_MC2_155 VASWRDHV-GGQVRVEHWSNVADQARAMVP---------ALLGQEASA-V
TH_3739|M.thermoresistible__bu VASWRNTV-GKQVRVEHWSNVADQARVLVP---------ALLGQEVPV-A
MAB_3838c|M.abscessus_ATCC_199 VAAWAGAD-GRPNRVEHWSNVADQVRALVP---------ALLGQETSANT
MLBr_02387|M.leprae_Br4923 VTGKLQLAHVAEAQGVVAAEAIAGAETLALSDYRMMPRATFCQPNVASFG
*: : ::. . :. : :
Mvan_0467|M.vanbaalenii_PYR-1 EPVEYVWSDQYDWKIQFCGQTTHLGEPLVVGDRLSDNRFTALYPDREQRL
Mflv_0680|M.gilvum_PYR-GCK EPVEYVWSDQYDWKIQFCGQTTHLGEPLVVGDRLSDNRFTVLYPDGEQRL
Mb0707|M.bovis_AF2122/97 VVVPYFWSDQYDVKIQCLGEPHATDVVHLVED---DGRKFLAYYERDGVL
Rv0688|M.tuberculosis_H37Rv VVVPYFWSDQYDVKIQCLGEPHATDVVHLVED---DGRKFLAYYERDGVL
MMAR_1017|M.marinum_M VVVPYFWSDQYDVKIQCLGEPHSTDVVHLVED---DGRKFLAYYERDGVL
MUL_0769|M.ulcerans_Agy99 VVVPYFWSDQYDVKIQCLGEPHSTDVVHLVED---DGRKLLAYYQRDGVL
MAV_4485|M.avium_104 VVVPYFWSDQYDVKIQCLGEPEADDIVHIVED---DGRKFLAYYERDGAL
MSMEG_1416|M.smegmatis_MC2_155 VSVPYFWSDQYDVKIQCLGEPESDDTVHIVED---DGRKFLAYYERDGVL
TH_3739|M.thermoresistible__bu ATVPYFWSDQYDVKIQCLGEPEAGDTVHVVSD---DGRKFLAYYERDGVL
MAB_3838c|M.abscessus_ATCC_199 VAVPYFWSDQYDVKIQSLGHLGSSDTVHLISD---DGRKFLAYLERDGVL
MLBr_02387|M.leprae_Br4923 LTEQQARDGGYDVVVAKFPFTANAKAHGMGDP---SGFVKLVADAKYGEL
.. ** : : .. *
Mvan_0467|M.vanbaalenii_PYR-1 SG------AAIVNWPRALIECRRALKAGSDLETVRERLELLTNPPDTAAT
Mflv_0680|M.gilvum_PYR-GCK GG------AAVVNWPRALIECRRALQAGSDLESVRGRLEVLTNPPDTAAT
Mb0707|M.bovis_AF2122/97 VG------VVGGGMAGKVMKVRGKIAAGAPIAEVLDQTQA----------
Rv0688|M.tuberculosis_H37Rv VG------VVGGGMAGKVMKVRGKIAAGAPIAEVLDQTQA----------
MMAR_1017|M.marinum_M VG------VVGGGVPGKVMKVRAKIAAAVPISEMLDESQG----------
MUL_0769|M.ulcerans_Agy99 VG------VVGGGVPGKVMKVRAKIAAAVPISEMLDESQGQTSARIPARG
MAV_4485|M.avium_104 VG------VVGGGMPGKVMKTRAKIAAAVPIAEMLG--------------
MSMEG_1416|M.smegmatis_MC2_155 VG------VVGGGMPGKVMKTRGKIAAGAPIAEVLGG-------------
TH_3739|M.thermoresistible__bu TG------VVGGGMPGKVMKARAKIAAGAPISEVLE--------------
MAB_3838c|M.abscessus_ATCC_199 TG------VVGCGMAGPVMKMRAKIAAGTPIAEVLAGS------------
MLBr_02387|M.leprae_Br4923 LGGHMIGHNVSELLPELTLAQKWDLTATELVRNVHTHPTLSEALQECFHG
* . . : : : * : :
Mvan_0467|M.vanbaalenii_PYR-1 RSP-----------------------------------------------
Mflv_0680|M.gilvum_PYR-GCK RSP-----------------------------------------------
Mb0707|M.bovis_AF2122/97 --------------------------------------------------
Rv0688|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1017|M.marinum_M --------------------------------------------------
MUL_0769|M.ulcerans_Agy99 AGPGGPLVRWPTLTKSGRWRAEKATASSPVARVDGIGLEPGPRRTASWPD
MAV_4485|M.avium_104 --------------------------------------------------
MSMEG_1416|M.smegmatis_MC2_155 --------------------------------------------------
TH_3739|M.thermoresistible__bu --------------------------------------------------
MAB_3838c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02387|M.leprae_Br4923 LIGHMINF------------------------------------------
Mvan_0467|M.vanbaalenii_PYR-1 ----
Mflv_0680|M.gilvum_PYR-GCK ----
Mb0707|M.bovis_AF2122/97 ----
Rv0688|M.tuberculosis_H37Rv ----
MMAR_1017|M.marinum_M ----
MUL_0769|M.ulcerans_Agy99 VPGT
MAV_4485|M.avium_104 ----
MSMEG_1416|M.smegmatis_MC2_155 ----
TH_3739|M.thermoresistible__bu ----
MAB_3838c|M.abscessus_ATCC_199 ----
MLBr_02387|M.leprae_Br4923 ----