For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDSRWTVLIGAGFVAATMSGLVVLGVPTAFAAPGETETVSATESDTPSDAAPAETSAPDTEHEDEQSAPE HGIDAANPPLEADLEADLEAEYPDRERNGDKVIPVEGDVTDGAVGDEGAVGDEGVVGEDVVGDEDVDVDE DIAEPAEPAVIVESDAGITPATVTFTEEPRTDKDSPQRSLAETESVPLGATAETTPSRAPSLLNLVGSVV LNVLVGFIQLIDGPPVLPANSSVTVRTSTLNLPIGSGRSVQADWYFPETVDESTRLVYFQHGFLASGPMY SYTAARLAERTNAIVVAPSLTSNFFAPDAAWVGGSTMHRAVAELFVGDRAALAESAGAAAGHTITLPDKF VLVGHSAGGTLVTSAAGFMVDNGAIEHLAGVVMLDGVEPAGSRLVGDALAKLRGAHDRPIYLISSERYFW SRGGDMADKLQLARPDRFTGVGLKGGLHIDYMEGGNPIVQFGEYLVGGFSLPRNIDAAADITVGWVNDLF AGTMTEGVYGAPGQHIPVVTPSGTATAVVLPLGVPARPVWPPLLDTILTAVFDFGGRYLFVYEPLRGYQT AANPATSQQGALVGASRH
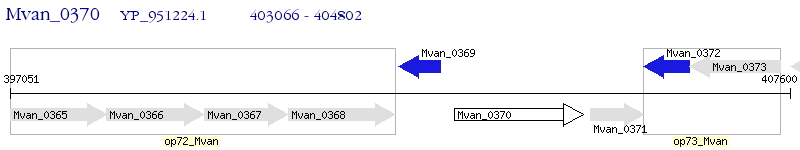
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0370 | - | - | 100% (578) | hypothetical protein Mvan_0370 |
| M. vanbaalenii PYR-1 | Mvan_1256 | - | 7e-99 | 41.85% (540) | hypothetical protein Mvan_1256 |
| M. vanbaalenii PYR-1 | Mvan_1255 | - | 5e-68 | 36.86% (529) | hypothetical protein Mvan_1255 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0365 | - | 1e-179 | 60.49% (567) | hypothetical protein Mflv_0365 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3402 | - | 1e-46 | 38.20% (322) | PE-PGRS family protein |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3222 | - | 6e-07 | 29.80% (198) | prolipoprotein diacylglyceryl transferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0370|M.vanbaalenii_PYR-1 MDSRWTVLIGAGFVAATMSGLVVLGVPTAFAAPGETETVSATESDTPSDA
Mflv_0365|M.gilvum_PYR-GCK MAHRRARLLLSASAAAAISGLLLIAAPAVSAEP----DGGGTETTVRSSA
MMAR_3402|M.marinum_M -----------------MSTYVIAAPDAFSVASGELAGIEAAVKGAAAAA
MSMEG_3222|M.smegmatis_MC2_155 -----MTTTVLAYLPSPSQGVWHLGPVPIRAYALCIIVGIVAALVIGDRR
. . . . . :
Mvan_0370|M.vanbaalenii_PYR-1 APAETSAPDTEHEDEQSAPEHGIDAANPPLEADLEADLEAEYPDRERNGD
Mflv_0365|M.gilvum_PYR-GCK KPADGDDRAAEADD-ASGPER------TDRKAGSDTDEAPRKP-RKPQGD
MMAR_3402|M.marinum_M APSTTGIISAAADEVSTAIAR----------------------LFGSYAD
MSMEG_3222|M.smegmatis_MC2_155 WQARGGEPGVIYDIALWAVPFG-------LAGGRIYHVITDWKTYFGPTG
: . . : . .
Mvan_0370|M.vanbaalenii_PYR-1 KVIPVEGDVTDGAVGDEGAVGDEGVVGEDVVGDEDVDVDEDIAEPAEPAV
Mflv_0365|M.gilvum_PYR-GCK RMILDS---PDAEAAEDPETPEEAADTERALT-ETTQIDPLPEAPPETST
MMAR_3402|M.marinum_M EFQTLSAQATRFQAQFERALSEAGAAYAAAEAANVSPLRALLQEAESLGV
MSMEG_3222|M.smegmatis_MC2_155 KGFGAALQIWEGGLGIWGAVAFGAVGAWIACRLRGIPLPAFGDAIAPGII
. .. . . :
Mvan_0370|M.vanbaalenii_PYR-1 IVESDAGIT--PAT--VTFTEEPRTDKDSPQRSLAETESVPLGATAETTP
Mflv_0365|M.gilvum_PYR-GCK VVPAERSES--TPTDPVPDTDIDTTAPGAAIVPIATVEATEANRASEAG-
MMAR_3402|M.marinum_M FAPIERLIG--RPLFGNGSVAAASTGATEASSLLTSGTNQAGLGGAVNYA
MSMEG_3222|M.smegmatis_MC2_155 LAQGIGRLGNYFNQELYGRPTDVPWGLEIYERLNKFGQSDQLNGVSTGQV
.. :
Mvan_0370|M.vanbaalenii_PYR-1 SRAPSLLNLVGSVVLNVLVGFIQLID-----GPPVLPANSSVTVRTSTLN
Mflv_0365|M.gilvum_PYR-GCK DRRPTLLNVVGSLVLNLLVGFIQVID-----GPPVLPPNSTVTVRTSSLT
MMAR_3402|M.marinum_M AAEALAAPATGATGVKSGFSFLQIPI-----GPSEFLG-----ITIPQFN
MSMEG_3222|M.smegmatis_MC2_155 TAVVHPTFLYELIWNIAVFGFLIWVDRKFRIGHGRLFALYVASYCVGRFW
..*: * : :
Mvan_0370|M.vanbaalenii_PYR-1 LPIGSGRSVQADWYFPET-----VDESTRLVYFQHG------------FL
Mflv_0365|M.gilvum_PYR-GCK LPIGGGRSVQADWYFPEEYSSGQLDSATRLVYLQHG------------FL
MMAR_3402|M.marinum_M FPAP------AHWYFPTQADG--SVNANGVIYLQHG------------FG
MSMEG_3222|M.smegmatis_MC2_155 VELMRSDTATEFAGIRVNTFTSTFVFIGAVVYIMLAPKGREEPESLRGKA
. : ::*: .
Mvan_0370|M.vanbaalenii_PYR-1 ASGPMYSYTAARLAERTNAIVVAPSLTSNFFAPDAAWVGGSTMHRAVAEL
Mflv_0365|M.gilvum_PYR-GCK ASGPLYSYTAARLAERTGAIVVAPSLTSNFFAADAAWVGGSTMHRAVADL
MMAR_3402|M.marinum_M AIGWFYQPLAMQLAQQTNSIVVVPTVPS-IPLPFGMWIGGAQMQQGVGSL
MSMEG_3222|M.smegmatis_MC2_155 ADETEEGDEESLVDEAGKELVAAAAGTGVVAAATAAAREDTDDTDGTTDP
* : : :*...: .. . . . .: .. .
Mvan_0370|M.vanbaalenii_PYR-1 FVGDRA----ALAESAGAAAGHTITLPDKFVLVGHSAGGTLVTSAAGFMV
Mflv_0365|M.gilvum_PYR-GCK FAGDRP----ALTQSASAAAGYVVRLPEKFVLVGHSAGGSLVTSAAGFMA
MMAR_3402|M.marinum_M FLGSQT----ALNISANQAG-YLGTLPQDFILSGHSAGGGLASIAAGSYL
MSMEG_3222|M.smegmatis_MC2_155 AAASDEPGDEAEPETEAATAVLTADGAEAMSVDEQPDDDEAVDEEAGQEA
.. * : :. .: : : :. .. . **
Mvan_0370|M.vanbaalenii_PYR-1 DNG--AIEHLAG-------------VVMLDGVEPAGSRLVGDALAKLRGA
Mflv_0365|M.gilvum_PYR-GCK DNG--AIDALTG-------------IVMLDGVEPAGSRLVGSALAKLRGS
MMAR_3402|M.marinum_M ANLGAATNHLQG-------------VVMFDGVASN-SSAFGAAVANLQAA
MSMEG_3222|M.smegmatis_MC2_155 EEPADELVELADEPEGEPEPAAAEEAEPAETIEPDEDEIFDAELAEALAE
: * . : : . . .. :*: .
Mvan_0370|M.vanbaalenii_PYR-1 HDRPIYLISSERYFWSRGGDMADKLQLAR--PDRFTGVGLKGGLHIDYME
Mflv_0365|M.gilvum_PYR-GCK SDRPVYLISSERYFWSRGGDMADKLQLAR--PGRFNGVGLQGGLHIDYME
MMAR_3402|M.marinum_M N-VPIYVVAAPPQPWNAFGATTNQLVSLY--PGQFTGVEIVNGSHVDSML
MSMEG_3222|M.smegmatis_MC2_155 AAEDFAVVPESAGSDEAEVAAAESAADAEDGPSDAESAEVPAEAAADDAV
. ::. . ::. *. .. : *
Mvan_0370|M.vanbaalenii_PYR-1 GGNPIVQFGEYLVGGFSLPRNIDAAADITVGWVNDLFAGTMTEG-VYGAP
Mflv_0365|M.gilvum_PYR-GCK GGNPLVQFGEYVVAGFSLRRNVDAASEITIGWVDDLFAGTAAG--VYGSP
MMAR_3402|M.marinum_M GGNPLIDFAAQLLTGFSPPGATQAVYTLSSGWINDIWAGADPLNPIYGIY
MSMEG_3222|M.smegmatis_MC2_155 VSEPAGTVDETEVAEEAAAEAADLEFESYDAELAEALAEAAEDMAVVVAP
.:* . : : : . : : * : :
Mvan_0370|M.vanbaalenii_PYR-1 GQHIPVVTPSGTATAVVLPLGVPARPVWPPLLDTILTAVFDFGGRYLFVY
Mflv_0365|M.gilvum_PYR-GCK GDVVAVVTPSGTATAVVLPLGEPARPVWPPLLDAALTAVFDFAGRHLFVY
MMAR_3402|M.marinum_M GPTGGYVAPGG----QAIFLGPTAGIVLPA--------------------
MSMEG_3222|M.smegmatis_MC2_155 AETAESDEAREPVETDATESEEPSEPEEPDAPEAVEAPEALDENADETRA
. . . .: *
Mvan_0370|M.vanbaalenii_PYR-1 EPLRGYQTAANPATSQQGALVGASRH--
Mflv_0365|M.gilvum_PYR-GCK EPLRGYDLQPTG----------------
MMAR_3402|M.marinum_M ----------------------------
MSMEG_3222|M.smegmatis_MC2_155 EPAAPAAVATAPVEPEKGRLRRWLRRNR