For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKLATAATKPLRGVGGFFAMSIDTFVAMVRPPFAFREYVLQSWFVARVSVLPALMLTLPYSVLLVFTFNI LLKEFGAADFAGTGAAIGTVSQIGPIVTVLVVSGAGATAMCADLGARTIREELDAMKVMGINPIQALVVP RVLAATTVALALSATVIIAGLAGAFFFCVFVQNVSAGAFISGMTLLTGAADVFVSLAKATLFGLAAGLIA CYKGISVGGGPAGVGNAVNETVVFTFMVLFAINVIATAVGIQFTVQ
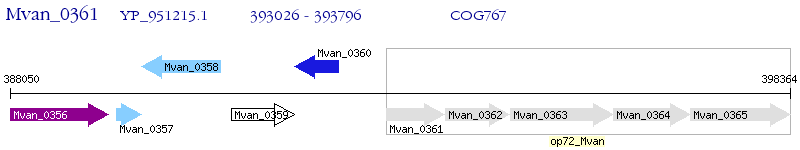
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0361 | - | - | 100% (256) | hypothetical protein Mvan_0361 |
| M. vanbaalenii PYR-1 | Mvan_3984 | - | e-120 | 86.75% (249) | hypothetical protein Mvan_3984 |
| M. vanbaalenii PYR-1 | Mvan_4606 | - | e-104 | 73.79% (248) | hypothetical protein Mvan_4606 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3531c | yrbE4A | 2e-86 | 60.32% (247) | integral membrane protein YrbE4a |
| M. gilvum PYR-GCK | Mflv_2099 | - | 1e-112 | 78.14% (247) | hypothetical protein Mflv_2099 |
| M. tuberculosis H37Rv | Rv1964 | yrbE3A | 9e-96 | 66.93% (254) | integral membrane protein YrbE3A |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 2e-78 | 55.02% (249) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_4155c | - | 2e-83 | 58.70% (247) | hypothetical protein MAB_4155c |
| M. marinum M | MMAR_4049 | yrbE3A_1 | 1e-109 | 76.00% (250) | integral membrane protein YrbE3A_1 |
| M. avium 104 | MAV_2059 | - | 1e-92 | 62.85% (253) | TrnB1 protein |
| M. smegmatis MC2 155 | MSMEG_0344 | - | 1e-111 | 80.32% (249) | hypothetical protein MSMEG_0344 |
| M. thermoresistible (build 8) | TH_0179 | - | 1e-119 | 84.68% (248) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE4A |
| M. ulcerans Agy99 | MUL_3916 | yrbE3A_1 | 1e-109 | 76.00% (250) | integral membrane protein YrbE3A_1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0361|M.vanbaalenii_PYR-1 MKLA----------TAATKPLRGVGGFFAMSIDTFVAMVRPPFAFREYVL
TH_0179|M.thermoresistible__bu MVLAP---------QPVTEPVRAVGGFFAMSLDTFVQMFRPPFAWREYVH
Rv1964|M.tuberculosis_H37Rv MVIVADK-AAGRVADPVLRPVGALGDFFAMTLDTSVCMFKPPFAWREYLL
MMAR_4049|M.marinum_M MVAPV----------AVAKPLRAFGGFYSMALDTFVAMVHPPFAWREYIA
MUL_3916|M.ulcerans_Agy99 MVTPV----------AVAKPLRAFGGFYSMALDTFVAMVHPPFAWREYIA
Mflv_2099|M.gilvum_PYR-GCK MVASG----------VIAKPMRAFGGFFSMALDTFVAMVKPPFAWREFIS
MSMEG_0344|M.smegmatis_MC2_155 MIALG----------PAAKPVGALGGFFAMTLDVFITIARPPFAWREYLN
MAV_2059|M.avium_104 MNATA-----------IAKPMTALGQFFLLSAEALAAAVRGPWAWREILE
Mb3531c|M.bovis_AF2122/97 ------------MIQQLAVPARAVGGFFEMSMDTARAAFRRPFQFREFLD
MAB_4155c|M.abscessus_ATCC_199 ------------MKQQLAAPARAVGGFVSMSLATFRNIFRRPFQGQEFLD
MLBr_02587|M.leprae_Br4923 MTTETRSSLVEYAHTKLQTPLVLVGGFFRMLVLTGKALFRRPFQWREFIL
* .* * : . : *: :* :
Mvan_0361|M.vanbaalenii_PYR-1 QSWFVARVSVLPALMLTLPYSVLLVFTFNILLKEFGAADFAGTGAAIGTV
TH_0179|M.thermoresistible__bu QSWFVARVSVVPALLLTMPYSVLLVFTFNVLLAEFGAADYSGTGAAIGTV
Rv1964|M.tuberculosis_H37Rv QCWFVARVSTLPGVLMTIPWAVISGFLFNVLLTDIGAADFSGTGCAIFTV
MMAR_4049|M.marinum_M QAWFVARVSLLPTLMLTIPYTVLLTFIENILLTEIGAADFSGTGAALGSV
MUL_3916|M.ulcerans_Agy99 QAWFVARVSLLPTLMLTIPYTVLLTFIENILLTEIGAADFSGTGAALGSV
Mflv_2099|M.gilvum_PYR-GCK QSWFVARVSIVPTLMLTIPYTVLLTFTFNILLTEFGAADFSGTGAALGTV
MSMEG_0344|M.smegmatis_MC2_155 QTWFVARVSLVPALMLTVPYTVLLVFTFNILLIEFGAADFSGTGAAIGTV
MAV_2059|M.avium_104 QIWFVARVSIFPTIMLSIPYTVLIVFVLNILLVEIGAGDLSGAGAGLASV
Mb3531c|M.bovis_AF2122/97 QTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGAADLSGAGTAFGTI
MAB_4155c|M.abscessus_ATCC_199 QTWFIARVSLLPTLLVAIPFTVLVAFTLNILLREIGAADLSGAATAFGTV
MLBr_02587|M.leprae_Br4923 QCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSAADLSGAGAAIGAV
* *:: **: .* :::::* :*: * *:** ::.*.* :*:. .: ::
Mvan_0361|M.vanbaalenii_PYR-1 SQIGPIVTVLVVSGAGATAMCADLGARTIREELDAMKVMGINPIQALVVP
TH_0179|M.thermoresistible__bu NQIGPIVTVLVVSGAGATAMCADLGARTIREELDALRVMGINPIQALVVP
Rv1964|M.tuberculosis_H37Rv NQSAPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGINPIQALAAP
MMAR_4049|M.marinum_M RQIGPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGVDPIQALVVP
MUL_3916|M.ulcerans_Agy99 RQIGPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGVDPIQALVVP
Mflv_2099|M.gilvum_PYR-GCK RQIGPIVTVLVVAGAGATAMCADLGARTIREELDALRVMGVNPIQALVVP
MSMEG_0344|M.smegmatis_MC2_155 SQIGPIVTVLVVAGAGSTAMCADLGARTIREELDAMRVMGIDPIQALVVP
MAV_2059|M.avium_104 TQVGPVVTAMVVSGAGSTAMCADLGARTIREEIDAMKVIGVNPVQALVVP
Mb3531c|M.bovis_AF2122/97 TQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMRVLGIDPIQRLVVP
MAB_4155c|M.abscessus_ATCC_199 TQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMQVLGIDPIQRLVVP
MLBr_02587|M.leprae_Br4923 TQLGPLTTVLVVAGAGSTSICADLGARTIREEIDAMEVLGIDPIHRLVVP
* .*:.*.:**:***:*::************:**:.*:*::*:: *..*
Mvan_0361|M.vanbaalenii_PYR-1 RVLAATTVALALSATVIIAGLAGAFFFCVFVQNVSAGAFISGMTLLTGAA
TH_0179|M.thermoresistible__bu RVLAATTVSLALASTVILVGLVGAFFFVVYVQHVSAGAFVTGLTLMTGAA
Rv1964|M.tuberculosis_H37Rv RVLAATTVSLALNSVVTATGLIGAFFCSVFLMHVSAGAWVTGLTTLTHTV
MMAR_4049|M.marinum_M RVLAATTVSLALSASVILVGLSGAYFFCVFIQHVSPGAFAAGLTLIIGTT
MUL_3916|M.ulcerans_Agy99 RVLAATTVSLALSASVILVGLSGAYFFCVFIQHVSPGAFAAGLTLIIGTT
Mflv_2099|M.gilvum_PYR-GCK RVLAATLVSLALSATVILVGLAGAYFFCVYIQNVSPGAFAAGLTLIIGAT
MSMEG_0344|M.smegmatis_MC2_155 RVLAATTVALALSATVIIVGLAGGFVFAVFIQHVPPGSFVAGLTVLTHGA
MAV_2059|M.avium_104 RIIAATFVAVMLYAVVAVTGLTGSYIFVVFVQHVTPGAFVAGMTLVTGLP
Mb3531c|M.bovis_AF2122/97 RVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPGAFINGLTVLTGLR
MAB_4155c|M.abscessus_ATCC_199 RVLASTFVALLLNGLVCAIGMVGGYVFSVFLQGVNPGAFINGLTVLTGLG
MLBr_02587|M.leprae_Br4923 RVLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGGAYLATLTTITGLP
*::*:* *: * . * *: *.: *:: * *:: :* :
Mvan_0361|M.vanbaalenii_PYR-1 DVFVSLAKATLFGLAAGLIACYKGISVGGGPAGVGNAVNETVVFTFMVLF
TH_0179|M.thermoresistible__bu DVIVSLVKATLFGMAAGLIACYKGISVGGGPAGVGNAVNETVVFSFMVLF
Rv1964|M.tuberculosis_H37Rv DVVISMIKATLFGLMAGLIACYKGMSVGGGPAGVGRAVNETVVFAFIVLF
MMAR_4049|M.marinum_M DVVIALVKAALFGLSAGLIACYKGISVGGGPAGVGNAVNETVVFTFMALF
MUL_3916|M.ulcerans_Agy99 DVVIALVKAALFGLSAGLIACYKGISVGGGPAGVGNAVNETVVFTFMALF
Mflv_2099|M.gilvum_PYR-GCK DVIIALVKAALFGLSAGMIACYKGISVGGGPAGVGNAVNETVVFTFMALF
MSMEG_0344|M.smegmatis_MC2_155 DVAVALAKAALFGLSAGLIACYKGISVGGGPAGVGNAVNETVVFTFMALF
MAV_2059|M.avium_104 QVVISLIKATLFGLSAGLIACYKGLSVGGGPTGVGNAVNETVVFSFMALF
Mb3531c|M.bovis_AF2122/97 ELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNAVNETVVYAFICLF
MAB_4155c|M.abscessus_ATCC_199 ELMISEVKAFLFGIFAGLVGCYRGLTVKGGPKGVGDAVNETVVYAFICLF
MLBr_02587|M.leprae_Br4923 EVVIATVKAATFGLIAGLVGCYRGLIVRGGSNGLGAAVNETVVLCVVALY
:: :: ** **: **::.**:*: * **. *:* ******* .: *:
Mvan_0361|M.vanbaalenii_PYR-1 AINVIATAVGIQFTVQ-
TH_0179|M.thermoresistible__bu AINIVVTAVGIQFTV--
Rv1964|M.tuberculosis_H37Rv VINIVVTAVGIPFMVS-
MMAR_4049|M.marinum_M AINIVATAVAIKVTL--
MUL_3916|M.ulcerans_Agy99 AINIVATAVAIKVTL--
Mflv_2099|M.gilvum_PYR-GCK AINIVATAVAVKVTM--
MSMEG_0344|M.smegmatis_MC2_155 AINVIATAVGVKITVV-
MAV_2059|M.avium_104 FINILTTALGVKVTAK-
Mb3531c|M.bovis_AF2122/97 VINVVMTAIGVRISAQ-
MAB_4155c|M.abscessus_ATCC_199 VINVVMTAIGVRVLVKH
MLBr_02587|M.leprae_Br4923 AVNVVLTTIGVRFGTGH
:*:: *::.: .