For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGSIGTNRSTIKGVTMLEQDLTDEDFAPYFVKAQEVSQFFREIGPKYDQENTFAYPSIEVFKKSGLGAL PVPKAFGGPGGNILQVSKVVSELSKGDSAITLAYNMHYIMVGIAGNLMSEEQNKYWLGRVADGELMFGPF SEQRAGFSGLADMQAVPQKDGGWKLYGKKTWGTLCEAADIITTNATETDADGNLPDTFDGRVAAEKLFIG DFKVDENGVGDGIRIEKTWDALGMHATGTQTIVFDGYFVPEDGFVSVWRAGAFGVLEWASLMFASIYLGM QQRILEESVKSLSKKHLGATFGAIVAADTEVKSVGHIIDGIGDMASRVECSRRVLYQTCQDLLDGKDKLW APELRFPYLGLAKTFIAGNVMHMSRQAMSMVGGQSFRRGGIFERLYRDSAASMFQPLNGDQTRTYIGQFL LESAAAQNGASS
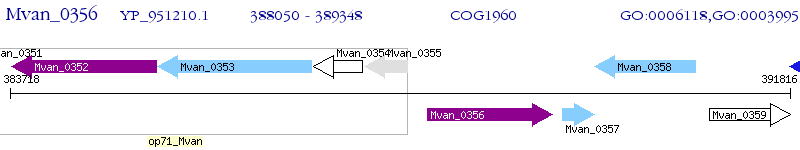
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0356 | - | - | 100% (432) | acyl-CoA dehydrogenase type 2 |
| M. vanbaalenii PYR-1 | Mvan_4089 | - | 9e-18 | 23.98% (367) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1974 | - | 7e-15 | 25.41% (429) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2528c | fadE19 | 4e-18 | 24.02% (408) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2554 | - | 3e-16 | 23.64% (368) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv2500c | fadE19 | 4e-18 | 24.02% (408) | acyl-CoA dehydrogenase |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-12 | 24.10% (390) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4538c | - | 9e-16 | 24.79% (363) | putative acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_3847 | fadE19 | 2e-18 | 23.37% (368) | acyl-CoA dehydrogenase FadE19 |
| M. avium 104 | MAV_1672 | - | 2e-19 | 24.32% (370) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0323 | - | 0.0 | 86.13% (411) | acyl-CoA dehydrogenase, short-chain specific, putative |
| M. thermoresistible (build 8) | TH_2782 | - | 3e-18 | 24.86% (370) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_3778 | fadE19 | 5e-18 | 23.10% (368) | acyl-CoA dehydrogenase FadE19 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3847|M.marinum_M -------------MTTAISAGTLPKEYQDLRDTVADFARSVVAPVSAKHD
MUL_3778|M.ulcerans_Agy99 -------------MTTAISAGTLPKEYQDLRDTVADFARSVVAPVSAKHD
MAV_1672|M.avium_104 -------------MTTSITAGTLPKEYQDLRDTVAEFARTVVAPVSAKHD
Mb2528c|M.bovis_AF2122/97 ----------MTTTTTTISGGILPKEYQDLRDTVADFARTVVAPVSAKHD
Rv2500c|M.tuberculosis_H37Rv ----------MTTTTTTISGGILPKEYQDLRDTVADFARTVVAPVSAKHD
Mflv_2554|M.gilvum_PYR-GCK -------------MSDFLSTGTLPDHYAQLAKTVRDFAQSVVAPVAAKHD
TH_2782|M.thermoresistible__bu -------------MPDYLTTGNLPDHYAELAGTVRDFAQSVVAPVAAKHD
MAB_4538c|M.abscessus_ATCC_199 ---------------MTFSTAELPDEYESLRKTVEEFAHSVVAPAAAHHD
MLBr_00737|M.leprae_Br4923 --------MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVD
Mvan_0356|M.vanbaalenii_PYR-1 MTGSIGTNRSTIKGVTMLEQDLTDEDFAPYFVKAQEVS-QFFREIGPKYD
MSMEG_0323|M.smegmatis_MC2_155 ----------------MLEQDMTDEEFEPYFAKAQEVS-QFFREIGPKYD
: ... . .: . ... *
MMAR_3847|M.marinum_M EEHSFPYEVVAKMGEMGLFGLPFPEEYGGMGGDYFALALALEELGKVDQS
MUL_3778|M.ulcerans_Agy99 EEHSFPYEVVAKMGEMGLFGLPFPEEYGGMGGDYFALALALEELGKVNQS
MAV_1672|M.avium_104 EEHSFPYEVVAKMGEMGLFGLPFPEEYGGMGGDYFALSLALEELGKVDQS
Mb2528c|M.bovis_AF2122/97 AEHSFPYEIVAKMGEMGLFGLPFPEEYGGMGGDYFALSLVLEELGKVDQS
Rv2500c|M.tuberculosis_H37Rv AEHSFPYEIVAKMGEMGLFGLPFPEEYGGMGGDYFALSLVLEELGKVDQS
Mflv_2554|M.gilvum_PYR-GCK EEHSFPYEVVSGMADMGLFGLPFPEEYGGMGGDYFALCLALEELGKVDQS
TH_2782|M.thermoresistible__bu QERSFPYEVVAAMGEMGLFGLPFPEEYGGMGGDYFALCLALEELGKVDQS
MAB_4538c|M.abscessus_ATCC_199 ATKTFPYAVVAQMGEMGLFGLPFPEEYGGMGGDYFALCLTLEELARADQS
MLBr_00737|M.leprae_Br4923 QRARFPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDAS
Mvan_0356|M.vanbaalenii_PYR-1 QENTFAYPSIEVFKKSGLGALPVPKAFGGPGGNILQVSKVVSELSKGDSA
MSMEG_0323|M.smegmatis_MC2_155 QENTFAYPSIEAFKKSGLGALPVPRAFGGPGGNILHVSKVVTELSRGDSA
*. : : *: .: .*. :** *.: . . .: *:.: : :
MMAR_3847|M.marinum_M VAITLEAGVGLGAMPIYRFGNEEQKSKWLPDLLAGRALAGFGLTEPGAGS
MUL_3778|M.ulcerans_Agy99 VAITLEAGVGLGAMPIYRFGNEEQKSKWLPDLLAGRALAGFGLTEPGAGS
MAV_1672|M.avium_104 VAITLEAGVSLGAMPIYRFGNEEQKRKWLPDLTAGRALAGFGLTEPGAGS
Mb2528c|M.bovis_AF2122/97 VAITLEAAVGLGAMPIYRFGTEEQKQKWLPDLTSGRALAGFGLTEPGAGS
Rv2500c|M.tuberculosis_H37Rv VAITLEAAVGLGAMPIYRFGTEEQKQKWLPDLTSGRALAGFGLTEPGAGS
Mflv_2554|M.gilvum_PYR-GCK VAITLEAGVSLGAMPVYRFGNESQKQEWLPLLASGKALGAFGLTEAGGGS
TH_2782|M.thermoresistible__bu VAITLEAGVSLGAMPIYRFGTEEQKQRWLPELAAGKALGAFGLTEPGGGS
MAB_4538c|M.abscessus_ATCC_199 VAITLEAAVSLGAMPVYRYGSEEQKQRWLPELASGKSLAAFGLTEPGGGT
MLBr_00737|M.leprae_Br4923 ASL-IPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGS
Mvan_0356|M.vanbaalenii_PYR-1 ITLAYNMHYIMVGIAGNLMS-EEQNKYWLGRVADGELMFGP-FSEQRAGF
MSMEG_0323|M.smegmatis_MC2_155 ITLAYNMHYIMLGIAGNLMS-EEQNKYWMGRVIDGELIFGP-FSEQRAGF
:: : : . *. : : : : . ::* .*
MMAR_3847|M.marinum_M D-AGSTRTTARLDGGEWVVNGSKQF--ITNSGTDITSLVTITAVTG---S
MUL_3778|M.ulcerans_Agy99 D-AGSTRTTARLDGGEWVVNGSKQF--ITNSGTDITSLVTITAVTG---S
MAV_1672|M.avium_104 D-AGATRTTARLEDGEWVINGSKQF--ITNSGTDITSLVTVTAVTG---T
Mb2528c|M.bovis_AF2122/97 D-AGSTRTTARLEGDEWIINGSKQF--ITNSGTDITSLVTVTAVTGTTGT
Rv2500c|M.tuberculosis_H37Rv D-AGSTRTTARLEGDEWIINGSKQF--ITNSGTDITSLVTVTAVTGTTGT
Mflv_2554|M.gilvum_PYR-GCK D-AGATKTTAKSDGASWVINGTKQF--ITNSGTDITKLVTVTAVTG----
TH_2782|M.thermoresistible__bu D-AGATRTTARLDDGHWVINGSKQF--ITNSGTDITKLVTVTAVTG----
MAB_4538c|M.abscessus_ATCC_199 DIPGSIRTRAVEDNGSWVINGAKAF--ITNSGTDITAVVGVAAVTG---R
MLBr_00737|M.leprae_Br4923 D-AASMRTRAKADGDDWILNGFKCW--ITNGGK--STWYTVMAVTD----
Mvan_0356|M.vanbaalenii_PYR-1 SGLADMQAVPQKDGG-WKLYGKKTWGTLCEAADIITTNATETDADGNLPD
MSMEG_0323|M.smegmatis_MC2_155 SGLADMKAVPQPGGG-WRLYGKKTWGTLCEAASIITTNATETDANGNLPE
. . :: . . * : * * : : :.. : . .
MMAR_3847|M.marinum_M IADGKKEISTIIVPSGTP-------GFIVEPVYNKVGWNASDTHPLSFDD
MUL_3778|M.ulcerans_Agy99 IADGKKEISTIIVPSGTP-------GFIVEPVYNKVGWNASDTHPLSFDD
MAV_1672|M.avium_104 VGADKKEISTIIVPSGTP-------GFTVEPVYSKVGWNASDTHPLSFSD
Mb2528c|M.bovis_AF2122/97 AADAKKEISTIIVPSGTP-------GFTVEPVYNKVGWNASDTHPLTFAD
Rv2500c|M.tuberculosis_H37Rv AADAKKEISTIIVPSGTP-------GFTVEPVYNKVGWNASDTHPLTFAD
Mflv_2554|M.gilvum_PYR-GCK ENNGKKEISSILVPVPTE-------GFTAEPAYNKVGWNASDTHPLSFDD
TH_2782|M.thermoresistible__bu EVAGKKEISAIIVPVPTD-------GFTVEPAYHKVGWNASDTHPLSFDD
MAB_4538c|M.abscessus_ATCC_199 SADGSPEISVIHVPSGTP-------GFTVEPGYDKVGWHASDTHPLSFDD
MLBr_00737|M.leprae_Br4923 PDKGANGISAFIVHKDDE-------GFSIGPKEKKLGIKGSPTTELYFDK
Mvan_0356|M.vanbaalenii_PYR-1 TFDGRVAAEKLFIGDFKVDENGVGDGIRIEKTWDALGMHATGTQTIVFDG
MSMEG_0323|M.smegmatis_MC2_155 TFEERVAAEKLFIGDFTVDENGAGDGIRIEKTWDALGMHATGTQTIVFDG
. : : *: :* :.: * : *
MMAR_3847|M.marinum_M ARVPEENLLGIRGKGYANFLSILDEGRIAIAALATGVAQGCVDESVKYAK
MUL_3778|M.ulcerans_Agy99 ARVPEENLLGIRGKGYANFLSILDEGRIAIAALATGVAQGCVDESVKYAK
MAV_1672|M.avium_104 ARVPEENLLGARGTGYANFLSILDEGRIAIAALATGAAQGCVDESVKYAK
Mb2528c|M.bovis_AF2122/97 ARVPRENLLGARGSGYANFLSILDEGRIAIAALATGAAQGCVDESVKYAN
Rv2500c|M.tuberculosis_H37Rv ARVPRENLLGARGSGYANFLSILDEGRIAIAALATGAAQGCVDESVKYAN
Mflv_2554|M.gilvum_PYR-GCK VRVPQENLLGERGRGYANFLRILDEGRIAIAALSVGAAQGCVDESVKYAK
TH_2782|M.thermoresistible__bu VRVPEENLLGERGRGYANFLRILDEGRIAIAALSVGAAQGCVDESVKYAK
MAB_4538c|M.abscessus_ATCC_199 VRVPYENLLGERGRGYAGFLRILDEGRIAIAAVATGAAQGCVDESLRYAR
MLBr_00737|M.leprae_Br4923 CRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTK
Mvan_0356|M.vanbaalenii_PYR-1 YFVPEDGFVSVWRAG---AFGVLEWASLMFASIYLGMQQRILEESVKSLS
MSMEG_0323|M.smegmatis_MC2_155 YFVPEDGFVSVWRAG---AFGTLEWASLMFASIYLGMQQRILEEATNALA
:* : ::. * : *: :.: * * :: :
MMAR_3847|M.marinum_M ERQS---FGQPIGSYQAISFK------IARMEARAHVARTAYYEAAAKML
MUL_3778|M.ulcerans_Agy99 ERQS---FGQPIGSYQAISFK------IARMEARAHVARTAYYEAAAKML
MAV_1672|M.avium_104 ERQS---FGQPIGSYQAISFK------IARMEARAHVARTAYYDAAAKML
Mb2528c|M.bovis_AF2122/97 QRQS---FGQPIGAYQAIGFK------IARMEARAHVARTAYYDAAAKML
Rv2500c|M.tuberculosis_H37Rv QRQS---FGQPIGAYQAIGFK------IARMEARAHVARTAYYDAAAKML
Mflv_2554|M.gilvum_PYR-GCK ERQA---FGAAIGTYQAIAFK------IARMEARAHTARAAYYDAAALML
TH_2782|M.thermoresistible__bu EREA---FGRPIGANQAIAFK------IARMEARAHTARTAYYDAAALML
MAB_4538c|M.abscessus_ATCC_199 SRPA---FGSAIIDYQSVSFK------IARMQARAHTARLAYYHAAALLL
MLBr_00737|M.leprae_Br4923 DRKQ---FGESISTFQSIQFM------LADMAMKVEAARLIVYAAAARAE
Mvan_0356|M.vanbaalenii_PYR-1 KKHLGATFGAIVAADTEVKSVGHIIDGIGDMASRVECSRRVLYQTCQDLL
MSMEG_0323|M.smegmatis_MC2_155 KKHLGATFGAIVAEDTAVKDVGHIVDGIGDFASRVEWSRRVLHRTCEDLL
.: ** : : :. : :.. :* : :.
MMAR_3847|M.marinum_M AGKP-------FKKEAAIAKMISSEAAMDNARDATQVHGGYGFMNEYPVA
MUL_3778|M.ulcerans_Agy99 AGKP-------FKKEAAIAKMISSEAAMDNARDAAQVHGGYGFMNEYPVA
MAV_1672|M.avium_104 AGKP-------FKKEAAIAKMIASEAAMDNARDATQIHGGYGFMNEYPVA
Mb2528c|M.bovis_AF2122/97 AGKP-------FKKEAAIAKMISSEAAMDNSRDATQIHGGYGFMNEYPVA
Rv2500c|M.tuberculosis_H37Rv AGKP-------FKKEAAIAKMISSEAAMDNSRDATQIHGGYGFMNEYPVA
Mflv_2554|M.gilvum_PYR-GCK SGKP-------FKKAASVAKMVASEAAMDNARDATQIFGGYGFMNEYSVA
TH_2782|M.thermoresistible__bu AGKP-------FKKQAAIAKMVASEAAMDNARDATQIFGGYGFINEFPVA
MAB_4538c|M.abscessus_ATCC_199 AGKP-------FKTEASIAKLIASEAAMDNARDATQIFGGAGFMNESPVA
MLBr_00737|M.leprae_Br4923 RGEPD------LGFISAASKCFASDIAMEVTTDAVQLFGGAGYTSDFPVE
Mvan_0356|M.vanbaalenii_PYR-1 DGKDKLWAPELRFPYLGLAKTFIAGNVMHMSRQAMSMVGGQSFRRGGIFE
MSMEG_0323|M.smegmatis_MC2_155 EGRDKEWPAELRFPYLGLAKTFIADNVLHMSRHAMSMIGGSAFRKGTIFE
*. . :* . : .:. : .* .: ** .: .
MMAR_3847|M.marinum_M RHYRDSKILEIGEGTTEVQLMLIARSLGLS---------
MUL_3778|M.ulcerans_Agy99 RHYRDSKILEIGEGTTEVQLMLIARSLGLS---------
MAV_1672|M.avium_104 RHYRDSKILEIGEGTTEVQLMLIARSLGLS---------
Mb2528c|M.bovis_AF2122/97 RHYRDSKVLEIGEGTTEVQLMLIARSLGLQ---------
Rv2500c|M.tuberculosis_H37Rv RHYRDSKVLEIGEGTTEVQLMLIARSLGLQ---------
Mflv_2554|M.gilvum_PYR-GCK RHYRDSKILEIGEGTTEVQLMLIGRELGL----------
TH_2782|M.thermoresistible__bu RHYRDSKILEIGEGTTEVQLMLIGRELGL----------
MAB_4538c|M.abscessus_ATCC_199 RQYRDSKILEIGEGTSEVQLMLIARALTG----------
MLBr_00737|M.leprae_Br4923 RFMRDAKITQIYEGTNQIQRVVMSRALLR----------
Mvan_0356|M.vanbaalenii_PYR-1 RLYRDSAASMFQPLNGDQTRTYIGQFLLESAAAQNGASS
MSMEG_0323|M.smegmatis_MC2_155 RLYRDSAASMFQPLNADQTRTYVGQYLLEQAAAK-----
* **: : . : :.: *