For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTVFNVRNLKSPGASRATLIIGVIVVVLAVVGAVLGWNLYKQMTTKTVVAYFTQTLAIYPGDKVQIMGV QVGSIDKIEPAGDKMKVTFHYAGKYKVPQDATASILNPSLVASRTIQLSPPYTGGPVLADGAVLDVDRTQ VPVEYDDVRDSISRLLTDLGPTPEQPKGPFGDIIESAADGFAGKGEQLNKTLNNLSEALFALNEGRGDFF SVVKSLALFVNALHQSDQQFVALNNNLAQFTNAFTDTDREVADALRDLNELLTTTRNFVDENGEVLAHDV QNLADVTNAILQPEPRDGLETALHAYPNVAANLMNISSPNAGGIVTMPVISNFANPMQFLCSAIQAGSRL GYQESAELCAQYLGPILDAIKFNYLPFGANQLQTAMTLPKQIAYSEERLRPPPGYKDTTVPGIFSRDTLF SHGNHEPGWVVAPGMQGVEVQPFTQNMLTPESLAELMGGPDIVPPPAPPAFGTTRNGNLPGPPNAFDQNN PLPPPWYPQPGPPPAPAPGVVQGNPLGAIAPAAPPAGPAPAAPSGPLLPAEAGG
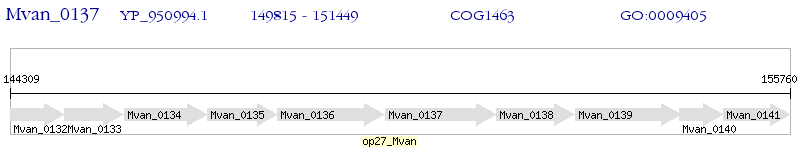
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0137 | - | - | 100% (544) | virulence factor Mce family protein |
| M. vanbaalenii PYR-1 | Mvan_4601 | - | 3e-81 | 41.86% (387) | virulence factor Mce family protein |
| M. vanbaalenii PYR-1 | Mvan_4147 | - | 1e-77 | 35.56% (540) | virulence factor Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0178 | mce1D | 0.0 | 63.44% (547) | MCE-family protein MCE1D |
| M. gilvum PYR-GCK | Mflv_0708 | - | 0.0 | 90.44% (544) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0592 | mce2D | 0.0 | 68.74% (515) | MCE-family protein MCE2D |
| M. leprae Br4923 | MLBr_02592 | mce1D | 0.0 | 60.58% (548) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4150c | - | 3e-68 | 33.89% (481) | MCE-family protein |
| M. marinum M | MMAR_0415 | mce1D | 0.0 | 60.81% (546) | MCE-family protein Mce1D |
| M. avium 104 | MAV_4548 | - | 0.0 | 67.40% (503) | virulence factor mce family protein |
| M. smegmatis MC2 155 | MSMEG_0137 | - | 0.0 | 77.47% (546) | virulence factor mce family protein |
| M. thermoresistible (build 8) | TH_3020 | - | 0.0 | 74.55% (440) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_1065 | mce1D | 0.0 | 60.81% (546) | MCE-family protein Mce1D |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0137|M.vanbaalenii_PYR-1 MSTVFNVRNLKSPGASRATLIIGVIVVVLAVVGAVLGWNLYKQMTTKTVV
Mflv_0708|M.gilvum_PYR-GCK MSTIFNVRNMKGPGLSRG-LIIGAVVLVLVVVGAFLGMKLYKQMTTKTVV
MSMEG_0137|M.smegmatis_MC2_155 MSTIFNIRNIQLPRLSRAAVIIGALVVAAALVAGYFGMNAYRKLTNTTVT
TH_3020|M.thermoresistible__bu MSTIFNVRNLRLPRLSRTAVILGALVVALALVAAVVGYQLYKRITTNTAV
MMAR_0415|M.marinum_M MSTIFDIRNIRLPKVSRATVIIGSLVLVLGLVAAFVGLKLYQKLTNNTVV
MUL_1065|M.ulcerans_Agy99 MSTIFDIRNIRLPEVSRATVIIGSLVLVLGLVAAFVGLKLYQKLTNNTVV
MLBr_02592|M.leprae_Br4923 MSTIFDVRKLKLLRLSRAPVIVGSLVVVLALVVGFVGWRLYQKLTNNTVV
Mb0178|M.bovis_AF2122/97 MSTIFDIRNLRLPQLSRASVVIGSLVVVLALAAGIVGVRLYQKLTNNTVV
Rv0592|M.tuberculosis_H37Rv MSTIFDIRSLRLPKLSAKVVVVGGLVVVLAVVAAAAGARLYRKLTTTTVV
MAV_4548|M.avium_104 MSTIFDIRNLRLPKVSARVLVIAALAAVFVFIAAVAGVQLYRKLTTTTVV
MAB_4150c|M.abscessus_ATCC_199 ------------MTTSKTWLKRGAFAMLSVALVGGLLYWVWPSRGTYKIV
* : . .. . : . . .
Mvan_0137|M.vanbaalenii_PYR-1 AYFTQTLAIYPGDKVQIMGVQVGSIDKIEPAGDKMKVTFHYAGKYKVPQD
Mflv_0708|M.gilvum_PYR-GCK AYFTQTLAIYPGDKVQIMGVQVGAIDKIEPAGDKMKVTFHYEDKYRVPEN
MSMEG_0137|M.smegmatis_MC2_155 AYFPEVLALYPGDKVLIMGVRVGSIDSIETAGDKMKVVFHFNNKYKVPEN
TH_3020|M.thermoresistible__bu AYFEQTLALYPGDKVQIMGVHVGNIDTIEPAGDKMKVTFHYDSKYKLPSD
MMAR_0415|M.marinum_M AYFPSANALYPGDKVQIMGLQVGKIDNIEPAGDKMKVTFHYQNKYKVPAN
MUL_1065|M.ulcerans_Agy99 AYFPSANALYPGDKVQIMGLQVGKIDNIEPAGDKMKVTFHYQNKYKVPAN
MLBr_02592|M.leprae_Br4923 VYFPAATALYTGDKVQIMGLRVGSIDSIVPAGDKMKVTFHYQNKYKVPAN
Mb0178|M.bovis_AF2122/97 AYFTQANALYVGDKVQIMGLPVGSIDKIEPAGDKMKVTFHYQNKYKVPAN
Rv0592|M.tuberculosis_H37Rv AYFSEALALYPGDKVQIMGVRVGSIDKIEPAGDKMRVTLHYSNKYQVPAT
MAV_4548|M.avium_104 AYFSETLALYPGDRVQIMGVRVGSIDKIEPAGDKMRVTLHYNNKYRVPAN
MAB_4150c|M.abscessus_ATCC_199 GHFASAVGLYPGDDVRILGVKAGRISAVEPREGDVKVTMEMPDKFKVPEG
:* . .:* ** * *:*: .* *. : . ..::*.:. .*:::*
Mvan_0137|M.vanbaalenii_PYR-1 ATASILNPSLVASRTIQLSPPYTGGPVLADGAVLDVDRTQVPVEYDDVRD
Mflv_0708|M.gilvum_PYR-GCK ATASILNPSLVASRTIQLSPAYTGGNTLPDNAVLDIDRTQVPVEYDDLRD
MSMEG_0137|M.smegmatis_MC2_155 ATASILNPSLVASRVIQLSPPYTGGPTLRDGAVLDVDRTQVPIEYDEVRN
TH_3020|M.thermoresistible__bu VSASILNPSLVASRSIQLSPPYTGGDTFPDGGVIPIERTQVPVEWDELRN
MMAR_0415|M.marinum_M ASAVILNPTLVASRTIQLEPPYKGGPVLADNAVIPLERTQVPVEWDELRN
MUL_1065|M.ulcerans_Agy99 ASAVILNPTLVASRTIQLEPPYKGGPVLADNAVIPLERTQVPVEWDELRN
MLBr_02592|M.leprae_Br4923 ASAVILNPTLVASRSIQLEPPYKGGPALGDNAVIPLERTQVPVEWDELRD
Mb0178|M.bovis_AF2122/97 ASAVILNPTLVASRNIQLEPPYRGGPVLADNAVIPVERTQVPTEWDELRD
Rv0592|M.tuberculosis_H37Rv ATASILNPSLVASRTIQLSPPYTGGPVLQDGAVIPIERTQVPVEWDQLRD
MAV_4548|M.avium_104 ATASILNPSLVASRTIQLSPPYTGGPVLRDGAVIPIERTRVPVEWDQLRD
MAB_4150c|M.abscessus_ATCC_199 AQAIIMAPNLVTARFVQLTPAYTDGKALQDGASLGLDKTAVPIEWDEVKT
. * *: *.**::* :** *.* .* .: *.. : :::* ** *:*:::
Mvan_0137|M.vanbaalenii_PYR-1 SISRLLTDLGPTPEQPKGPFGDIIESAADGFAGKGEQLNKTLNNLSEALF
Mflv_0708|M.gilvum_PYR-GCK SISRLLTDLGPTPEQPKGPFGDIIESAADGFAGKGKQLNSTLNNLSEALF
MSMEG_0137|M.smegmatis_MC2_155 QVTRLLADLGPTPEQPKGPFGDIIESFADGFAGKGEQLNRTLRGLSDALT
TH_3020|M.thermoresistible__bu SIDRILTDLGPTDEQPMGPFGDVVDSFADGLAGKGQQINQTLTSLSEALT
MMAR_0415|M.marinum_M SITNIIDKLGPTKDQPTGPFGEVLESFANGLEGKGKQINTTLDSLSRALT
MUL_1065|M.ulcerans_Agy99 SITNIIDKLGPTKDQPTGPFGEVLESFANGLEGKGKQINTTLDSLSRALI
MLBr_02592|M.leprae_Br4923 SITKIISKLGPTKEQPKGPFGDVIESFADGLAGKGKQINATLDSLSRALT
Mb0178|M.bovis_AF2122/97 SVSHIIDELGPTPEQPKGPFGEVIEAFADGLAGKGKQINTTLNSLSQALN
Rv0592|M.tuberculosis_H37Rv SINGILRQLGPTERQPKGPFGDLIESAADNLAGKGRQLNETLNSLSQALT
MAV_4548|M.avium_104 SINAILRQLGPTPQQPAGPFGDIIESAADNLSGKGKQVNETLNSLSQALT
MAB_4150c|M.abscessus_ATCC_199 ELAKLSDSLGPQ-GQSKGPVSDFINQAATTFDGNGDSFRNAVRELSQAAG
.: : .*** *. **..:.:: * : *:* ... :: ** *
Mvan_0137|M.vanbaalenii_PYR-1 ALNEGRGDFFSVVKSLALFVNALHQSDQQFVALNNNLAQFTNAFTDTDRE
Mflv_0708|M.gilvum_PYR-GCK ALNEGRGDFFSVVKSLATFVNALHQSDQQFVALNNDLAQFTNAFTDSDRE
MSMEG_0137|M.smegmatis_MC2_155 ALNEGRGDFFAVVKSLALFVNALHRSDQQFVALNNDLAQFTNSFTNTDQE
TH_3020|M.thermoresistible__bu ALNEGRGDFFGVVKSLALFVNALHRSDQQFVALNRDLAQFTTSFANTDQE
MMAR_0415|M.marinum_M ALNEGRGDFFAVVRSLALFVNALHQDDKQFVALNQNLADVTGRLVGYDRD
MUL_1065|M.ulcerans_Agy99 ALNEGRGDFFAVVRSLALFVNALHQDDKQFVALNQNLADVTGRLVGYDRD
MLBr_02592|M.leprae_Br4923 ALNEGRGDFFAVLRSLALFVNALHTDDQQFVALNQNLADFTTRLAHSDTD
Mb0178|M.bovis_AF2122/97 ALNEGRGDFFAVVRSLALFVNALHQDDQQFVALNKNLAEFTDRLTHSDAD
Rv0592|M.tuberculosis_H37Rv ALNEGRGDFVAITRSLALFVSALYQNDQQFVALNENLAEFTDWFTKSDHD
MAV_4548|M.avium_104 ALNEGRGDFVAITKSLARFVTALYQHKQQFVALNNNLAQFTDWFTQSDHQ
MAB_4150c|M.abscessus_ATCC_199 RLGDSRTDLFGTIKNLQTLVDALANSNEQIVQFSTHLASVSKVLAASTEN
*.:.* *:.. :.* :* ** .:*:* :. .**..: :. :
Mvan_0137|M.vanbaalenii_PYR-1 VADALRDLNELLTTTRNFVDENGEVLAHDVQNLADVTNAILQPEPRDGLE
Mflv_0708|M.gilvum_PYR-GCK VANALQDLDELLTTTRGFIDENGEVLAHDVQNLAEVTNAILQPEPLDGLE
MSMEG_0137|M.smegmatis_MC2_155 LANALQDLNRVLKTTREFLDRNGGVLTHDIDNLEQVTTAILQPEPRDGLE
TH_3020|M.thermoresistible__bu LANAVRDLDELLTTTRQFLDENAEVLTYDINNLAEVTNAILQPEPLDGLE
MMAR_0415|M.marinum_M LSNALQQFDSLLSTVRPFLDKNREVLTYDVNNLAEATNTLLQPDPLNGLE
MUL_1065|M.ulcerans_Agy99 LSNALQQFDSLLSTVRPFLDKNREVLTYDVNNLAEATNTLLQPDPLNGLE
MLBr_02592|M.leprae_Br4923 LSSALQQFNSMLSTVQSFLVKNREVLAADVNNLATVNNTVVQPEPLNGVE
Mb0178|M.bovis_AF2122/97 LSNAIQQFDSLLAVARPFFAKNREVLTHDVNNLATVTTTLLQPDPLDGLE
Rv0592|M.tuberculosis_H37Rv LADTVERIDDVLGTVRKFVSDNRSVLAADVNNLADATTTLVQPEPRDGLE
MAV_4548|M.avium_104 VSDTIAHLDQVLDAARKFVNDNGSALTHDVGNLADVTTTILQPEPRDGLE
MAB_4150c|M.abscessus_ATCC_199 LGATLGALNQALTDVRGFLGDTNSALIDQVNKLSDFVKIFS--DQSDDLE
:. :: :: * .: *. . .* :: :* . . : :.:*
Mvan_0137|M.vanbaalenii_PYR-1 TALHAYPNVAANLMNISSPNAGGIVTMPVIS---NFANPMQFLCS-AIQA
Mflv_0708|M.gilvum_PYR-GCK TALHAYPNVAANLMNISSPNAGGIVTMPVIS---NFANPMQFLCS-AIQA
MSMEG_0137|M.smegmatis_MC2_155 TGLHAYPNLAANVLNINSPNQGGIIGLPVLPGVTNFSNPLQFVCS-SIQA
TH_3020|M.thermoresistible__bu TALHVFPTMGANIMNISSPNSGGIVSLPVVT---NFANPLQFICS-AIQA
MMAR_0415|M.marinum_M TALHVLPTAASNVNQIYHPAHGSVVAVPEIT---SFANPMQFVCS-SIQA
MUL_1065|M.ulcerans_Agy99 TALHVLPTAASNVNHIYHPAHGSVVAVPEIT---SFANPMQFVCS-SIQA
MLBr_02592|M.leprae_Br4923 TFLHVAPTAAANMNQLYHPAHGSVVGVPTIT---NFANPMQFICS-AIQA
Mb0178|M.bovis_AF2122/97 TVLHIFPTLAANINQLYHPTHGGVVSLSAFT---NFANPMEFICS-SIQA
Rv0592|M.tuberculosis_H37Rv TALHVLPTYASNFNNLYYPLHSSLVGQFVFP---NFANPIQLICS-AIQA
MAV_4548|M.avium_104 TALHVLPTFAGNFNNLYQPAHSSLVGLFVFP---NFANPIHFLCS-AIQA
MAB_4150c|M.abscessus_ATCC_199 QILHVAPNGLANFYNIYNPAAGTLNGMLSIP---NLANPVQFICGGVFET
** *. .*. :: * . : .. .::**:.::*. :::
Mvan_0137|M.vanbaalenii_PYR-1 GSRLGYQESAELCAQYLGPILDAIKFNYLPFGANQLQTAMTLPKQIAYSE
Mflv_0708|M.gilvum_PYR-GCK GSRLGYQESAELCAQYLGPILDAIKFNYLPFGANQLQTAMTLPKQISYSE
MSMEG_0137|M.smegmatis_MC2_155 GSRLGYQESAELCAQYLAPIMDAIKFNYLPFGMNLASTAMTLPKQIAYSE
TH_3020|M.thermoresistible__bu GSRLGYQESAELCAEYLAPILDAIKFNFPPFGFNQITTASVLPKQVAYSE
MMAR_0415|M.marinum_M GSRLGYQESAELCAQYLAPVLDAIKFNYFPFGLNAFNTAEVLPKQVAYSE
MUL_1065|M.ulcerans_Agy99 GSRLGYQESAELCAQYLAPVLDAIKFNYFPFGLNAFNTAEVLPKQVAYSE
MLBr_02592|M.leprae_Br4923 GSRLGYQESAELCAQYLSPVLDAIKFNYFPFGLNLFSTAEVLPKEVAYSE
Mb0178|M.bovis_AF2122/97 GSRLGYQESAELCAQYLAPVLDAIKFNYFPFGLNVASTASTLPKEIAYSE
Rv0592|M.tuberculosis_H37Rv GSRLGYQESAELCAQYLAPVLDALKFNYLPFGSNPFSSAATLPKEVAYSE
MAV_4548|M.avium_104 GSRLGYQDSAELCAQYLAPVLDAIKFNYPPAGLNPFNSAATLPKEVAYSE
MAB_4150c|M.abscessus_ATCC_199 GTKTEYLKRAEICRERMGPVLRRATFNYAPILLHPINQISAYKGQIIYDT
*:: * . **:* : :.*:: .**: * : . :: *.
Mvan_0137|M.vanbaalenii_PYR-1 ERLRPPPGYKDTTVPGIFSRDTLFSHGNHEPGWVVAPGMQGVEVQPFTQN
Mflv_0708|M.gilvum_PYR-GCK ERLRPPPGYKDTTVPGIFSRDTLFSHGNHEPAWVVAPGMQGVEVQPFTRN
MSMEG_0137|M.smegmatis_MC2_155 KRLQPPPGYKDTTVPGIWSRDTLFSHGNHEPGWIVAPGMQGVQVQPATAN
TH_3020|M.thermoresistible__bu PRLQPPPGYKDTTVPGIWSRDTLFSHGNHEPGWIVAPGMQGIQVX-----
MMAR_0415|M.marinum_M PRLQPPNGYKDTTVPGIWVPDTPLSHRNTQPGWIVAPGMQGQQVGPITAG
MUL_1065|M.ulcerans_Agy99 PRLQPPNGYKDTTVPGIWVPDTPLSHRNTQPGWIVAPGMQGQQVGPITAG
MLBr_02592|M.leprae_Br4923 ARLQPPSGYKDTTVPGIWVPDTPLSHRNTQPGWIVAPGMQGQQVGPITAA
Mb0178|M.bovis_AF2122/97 PRLQPPNGYKDTTVPGIWVPDTPLSHRNTQPGWVVAPGMQGVQVGPITQG
Rv0592|M.tuberculosis_H37Rv ERLRPPPGYKDTTVPGIFSRDTPFSHGNHEPGWVVAPGMQGMQVQPFTAN
MAV_4548|M.avium_104 ERLRPPPGYKDTTVPGIFARDTPFSHGNHEPGWVVAPGMQGIDVQPFTAG
MAB_4150c|M.abscessus_ATCC_199 PETEAKSQAPREDLVWVRP------NGAPPPPKPSPQDLSALMLGPDEVN
. .. : : : * . .:.. :
Mvan_0137|M.vanbaalenii_PYR-1 MLTPESLAELMGGPDIVPPPAPPAFGTTRNGNLPGPPNAFDQNNPLPPPW
Mflv_0708|M.gilvum_PYR-GCK MLTPESLSELMGGPDVAPPPAPPAFGTTRNGNLPGPPNAFDQNNPLPPPW
MSMEG_0137|M.smegmatis_MC2_155 MLTPESLAELLGGPDIVPPPAPPAFGTTRGGNLPGPPNAFDENNPLPPPW
TH_3020|M.thermoresistible__bu ------------------XXAWDACRSTGSTSKIGSCSALRSRS------
MMAR_0415|M.marinum_M LMTPESLSELMGGPDIEPVQST-------LQTPPGPPNAYDEYPVLPPIG
MUL_1065|M.ulcerans_Agy99 LMTPESLSELMGGPDIEPVQST-------LQTPPGPPNAYDEYPVLPPIG
MLBr_02592|M.leprae_Br4923 LMTPDSLAELMGGPDIEPIQSN-------LQTPPGPPTAYDEHPVPPPIG
Mb0178|M.bovis_AF2122/97 LLTPESLAELMGGPDIAPPSSG-------LQTPPGPPNAYDEYPVLPPIG
Rv0592|M.tuberculosis_H37Rv MLTPESLAELLGGPDIAPPPPG-------TNLP-GPPNAYDES-------
MAV_4548|M.avium_104 MLTPESLAELMGGPDAAPPPPG-------VTAA-GPPNAYDES-------
MAB_4150c|M.abscessus_ATCC_199 APPPP-------GTEVRPAEPG--------PVPPGFPVTADDAP------
. * : .
Mvan_0137|M.vanbaalenii_PYR-1 YPQPGPP---PAPAPGVVQGNPLGAIAPAAPPAGPAPAAPSGPLLPAEAG
Mflv_0708|M.gilvum_PYR-GCK YPQPGPP---PAPAPGVVPGNPAPVNAPAP--AAPAPAAPSGPLLPAEAG
MSMEG_0137|M.smegmatis_MC2_155 YPQPGPP---PAPAPGVIPGDPLSAVAPAAP-AAPAAPAPAGPPLPAEAG
TH_3020|M.thermoresistible__bu --SSSAS---SSTVRVRLASNSSRCLVNR---------------------
MMAR_0415|M.marinum_M LQAP-VPIQPPPPGPEVIPGPVAPTPAPGPAPAPVG------APLPAEAG
MUL_1065|M.ulcerans_Agy99 LQAP-VPIQPPPPGPEVIPGPVAPTPAPGPAPAPVG------APLPAEAG
MLBr_02592|M.leprae_Br4923 LQAS-VPI--PSPRPDVVPGPVAPTPSP---AANVGD-----PPLPAELG
Mb0178|M.bovis_AF2122/97 LQAPQVPIPPPPPGPDVIPGPVPPTPAP------VG------APLPAEAG
Rv0592|M.tuberculosis_H37Rv ----------NPLPPPWYPQPASLPAAG----------------ATGQPG
MAV_4548|M.avium_104 ----------NPPSPPWFSRPT-------------------------PPG
MAB_4150c|M.abscessus_ATCC_199 --------------PPAIPGAPR---------------------------
Mvan_0137|M.vanbaalenii_PYR-1 G--
Mflv_0708|M.gilvum_PYR-GCK GTR
MSMEG_0137|M.smegmatis_MC2_155 AG-
TH_3020|M.thermoresistible__bu ---
MMAR_0415|M.marinum_M AGQ
MUL_1065|M.ulcerans_Agy99 AGQ
MLBr_02592|M.leprae_Br4923 AGQ
Mb0178|M.bovis_AF2122/97 GGQ
Rv0592|M.tuberculosis_H37Rv PGQ
MAV_4548|M.avium_104 PGR
MAB_4150c|M.abscessus_ATCC_199 ---