For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDTTTVLIVGAGFAGLGTAIRLLQQGIDDFVVLERADEVGGTWRDNTYPGAACDIPSLLYSYGFEQNPDW SRAYSGSAEILGYIKTMVDKYSLSRFIRFGVNVTGLEFDEESALWTAQTADGSQFTARTAVMASGPLANA SLPDIRGLDTFDGHKIHSARWDHDYDMSDKRVAVIGTGASAVQIIPELVKTARSVKVFQRTPGWVLPRPD FSHPQWVRTAFRRLPRVQNVGRQAWFWGHELMAVGMVWDTPVTTGIQLLAKANLRRQVSDTWLRRQLTPN FRAGCKRMLMSSDYYPALQRDNCKLVSWPIATLSPNGIRTADGIEHELDCIVFATGFDVCKAGTPFPIRG AHGRELSDEWSQGAYAYRSVTVAGYPNLFFTFGPNSGPGHNSALVYMEAEINYIVKAIGAIIANDLSTLE VREDRQNNYHGEVQQRLGSTTWNSGCKSWYLTNDGYNGTMYPGFATQFKRALSKVDMGDYVTTPTTARTE HHPTEHAQNGSNVAKVAQGRKAKVAQGRKSAGALPTRSDVKADILDVGVGKVPPKRAASRVRKDA*
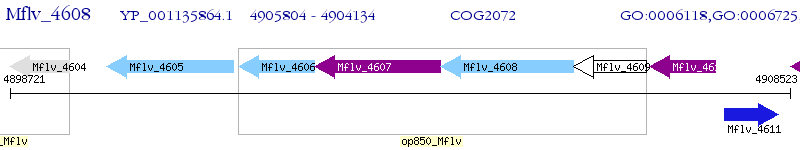
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4608 | - | - | 100% (556) | FAD dependent oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4287 | - | e-110 | 42.18% (486) | putative monooxygenase |
| M. gilvum PYR-GCK | Mflv_4605 | - | 8e-98 | 37.10% (496) | FAD dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3075c | - | 1e-117 | 44.10% (483) | monooxygenase |
| M. tuberculosis H37Rv | Rv3049c | - | 1e-117 | 44.10% (483) | monooxygenase |
| M. leprae Br4923 | MLBr_00065 | - | 6e-29 | 25.63% (437) | putative monooxygenase |
| M. abscessus ATCC 19977 | MAB_4476c | - | 0.0 | 75.21% (484) | putative monooxygenase |
| M. marinum M | MMAR_3139 | - | 0.0 | 72.02% (486) | monooxygenase |
| M. avium 104 | MAV_1795 | - | 0.0 | 62.73% (483) | flavin-binding monooxygenase |
| M. smegmatis MC2 155 | MSMEG_2310 | - | 1e-116 | 42.59% (479) | monooxygenase |
| M. thermoresistible (build 8) | TH_4080 | - | 1e-112 | 42.38% (479) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1880 | - | 1e-118 | 43.63% (479) | monooxygenase |
| M. vanbaalenii PYR-1 | Mvan_1519 | - | 1e-179 | 62.45% (466) | FAD dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3075c|M.bovis_AF2122/97 ---MSIADTAAKPSTPSPANQPPVRTRAVIIGTGFSGLGMAIALQKQGVD
Rv3049c|M.tuberculosis_H37Rv ---MSIADTAAKPSTPSPANQPPVRTRAVIIGTGFSGLGMAIALQKQGVD
MUL_1880|M.ulcerans_Agy99 MTDVVIPSTETTASGEPQAPTPPVKTRAVIIGTGFSGLGMAIALQKQGVD
TH_4080|M.thermoresistible__bu ---MTVTEPSADITPANTPTQPPVRTRALIVGTGFSGLGMAIALQRRGID
MSMEG_2310|M.smegmatis_MC2_155 ---------------MTAADQQPIRTRALVIGTGFSGLGMAIELQRRGVD
Mflv_4608|M.gilvum_PYR-GCK ---------------------MSDTTTVLIVGAGFAGLGTAIRLLQQGID
MAB_4476c|M.abscessus_ATCC_199 ------------------------MTEVLIVGAGFAGLGTAIRLLEKGIE
MMAR_3139|M.marinum_M ---------------------MTDTVTTLIVGAGFAGIGTAMRMLQAGVN
MAV_1795|M.avium_104 -------------------MARDHAHAALIIGAGFTGLGAAIRLAEAGVD
Mvan_1519|M.vanbaalenii_PYR-1 ----------------------MHVHDTLIVGAGFSGLGTAIKLRQAGVD
MLBr_00065|M.leprae_Br4923 ----------------MVVMAETEYLDVVIVGAGISGVSAAWHLQDRCPN
.:::*:*::*:. * : :
Mb3075c|M.bovis_AF2122/97 --FVILEKADDVGGTWRDNTYPGCACDIPSHLYSFSFEPKADWKHLFSYW
Rv3049c|M.tuberculosis_H37Rv --FVILEKADDVGGTWRDNTYPGCACDIPSHLYSFSFEPKADWKHLFSYW
MUL_1880|M.ulcerans_Agy99 --FVILEKSDDVGGTWRDNSYPGCACDIPSHLYSFSFEPKPDWRNPFSYQ
TH_4080|M.thermoresistible__bu --FLMLEKADDIGGTWRDNTYPGCACDIPSHLYSFSFEPKADWRHLFSYQ
MSMEG_2310|M.smegmatis_MC2_155 --FLILEKADEIGGTWRDNTYPGCACDIPSHMYSFSFEPKADWKHMWSFQ
Mflv_4608|M.gilvum_PYR-GCK D-FVVLERADEVGGTWRDNTYPGAACDIPSLLYSYGFEQNPDWSRAYSGS
MAB_4476c|M.abscessus_ATCC_199 D-FVILERGDDVGGTWRDNSYPGAACDIPSLLYSYSFEQNPQWSRAYSGG
MMAR_3139|M.marinum_M D-FVILERSHRVGGTWRDNTYPGAACDIPSLLYSYSFEPNPGWTRAYSGS
MAV_1795|M.avium_104 D-IVILERADRVGGTWRDTTYPGASCDVPSLLYSYSFVKNPTWSRTYSPA
Mvan_1519|M.vanbaalenii_PYR-1 D-IVIVERGQRVGGTWRDNTYPGVACDIPSLLYSFSFVRNPSWSRAYPTG
MLBr_00065|M.leprae_Br4923 KSYVILEKRAGMGGTWDLFRYPGIRSDSDMYTLGFRFRPWTKCQAIADGK
:::*: :**** *** .* .: * .
Mb3075c|M.bovis_AF2122/97 DEILGYLKGVTDKYGLRRYIEFNSLVDRGYWDDDECRWHVFTADG---RE
Rv3049c|M.tuberculosis_H37Rv DEILGYLKGVTDKYGLRRYIEFNSLVDRGYWDDDECRWHVFTADG---RE
MUL_1880|M.ulcerans_Agy99 PEIWDYLKGVTEKYGLRRYVEFNSLVDRAYWDDEECRWHVFTSDG---RE
TH_4080|M.thermoresistible__bu DEIQEYLQGVTAKYGLRRYIHFDSLVDRAHWDDTEHRWHVFTADG---RE
MSMEG_2310|M.smegmatis_MC2_155 PEIQDYLLGVTAKYGLRRHIHFNSHVDRAHWDDEELRWHVFTKTG---QE
Mflv_4608|M.gilvum_PYR-GCK AEILGYIKTMVDKYSLSRFIRFGVNVTGLEFDEESALWTAQTADG---SQ
MAB_4476c|M.abscessus_ATCC_199 SEILGYIQRMVDKHQLRRFIKFGVDVTGLTFDDSTGMWTADTAAG---EI
MMAR_3139|M.marinum_M AEILAYIDAIVAKYDLGRFIHFGADVSALEFDEDAGEWVVDTTGG---RR
MAV_1795|M.avium_104 PEIYRHLEDMADRFDIRRRIRFGHEVSGLAFDEDAGVWTATTKNR---KK
Mvan_1519|M.vanbaalenii_PYR-1 AEICDHIEDMADQHGLTPLIRFDTEVTGLTFDDQSGAWTVTTKRR---KQ
MLBr_00065|M.leprae_Br4923 P-ILEYIKSTAVMHGIDKYIRLNHKVTGADWSSIENRWTVQVENNGTPRM
* :: . . : :.:. * :.. * . .
Mb3075c|M.bovis_AF2122/97 YVAQFLISGAGALHIPS--FPEIAGRDEFAGPAFHSAQWDHSIDLTGKRV
Rv3049c|M.tuberculosis_H37Rv YVAQFLISGAGALHIPS--FPEIAGRDEFAGPAFHSAQWDHSIDLTGKRV
MUL_1880|M.ulcerans_Agy99 YVAQFLISGAGALHIPS--SPEIEGREEFAGPAFHSAEWDHSVDLTGKRV
TH_4080|M.thermoresistible__bu YIAQFLISCAGALHIPS--IPDFPGRDQFAGPAFHSAQWDHSVDLTGKRV
MSMEG_2310|M.smegmatis_MC2_155 YIAQFLVSGAGGLHIPQ--IPDIEGRESFAGAAFHSAQWDHSVDLRGKRV
Mflv_4608|M.gilvum_PYR-GCK FTARTAVMASGPLANAS--LPDIRGLDTFDGHKIHSARWDHDYDMSDKRV
MAB_4476c|M.abscessus_ATCC_199 FTGRCAVMAAGPLANVS--WPDIRGLDSYTGHKIHSARWDHDYEMSGKHV
MMAR_3139|M.marinum_M YRGRSVVMASGPLADAS--FPDIRGIESYEGKKIHSARWDHSYDMSGKRV
MAV_1795|M.avium_104 FRARTVVLASGPLSDVS--FPDIRGLDSYRGHKIHSARWDHDYDFAGKRV
Mvan_1519|M.vanbaalenii_PYR-1 FRARTVVLASGPLADHR--FPEIRGIDSYRGHKIHSASWDHDYDFTGKRV
MLBr_00065|M.leprae_Br4923 ISCSFLFLCSGYYNYEQGYAPTFLGSEDFTGPIIHPQHWPEDLDYAAKNI
. :* * : * : : * :*. * .. : *.:
Mb3075c|M.bovis_AF2122/97 AIVGTGASAIQIVPEIVGQ-VAELQLYQRTPPWVVPRTNEELPVSLRRAL
Rv3049c|M.tuberculosis_H37Rv AIVGTGASAIQIVPEIVGQ-VAELQLYQRTPPWVVPRTNEELPVSLRRAL
MUL_1880|M.ulcerans_Agy99 AIIGAGASAIQIVPEIVEQ-VGELQLYQRTPPWVVPRSNPEIPPALRRAM
TH_4080|M.thermoresistible__bu AVIGTGASAIQIVPEIIDR-VAQLQLYQRTPAWVVPRTNDELPAWLRRAL
MSMEG_2310|M.smegmatis_MC2_155 AVIGTGASAIQIVPEIVKD-VAELHLYQRTPAWVMPRVNNAFPQWLRNVF
Mflv_4608|M.gilvum_PYR-GCK AVIGTGASAVQIIPELVKT-ARSVKVFQRTPGWVLPRPDFSHPQWVRTAF
MAB_4476c|M.abscessus_ATCC_199 AVIGTGASGVQIIPELVKT-AARVKVFQRTPGWVLPRLDFQHPDWAQRVF
MMAR_3139|M.marinum_M AVIGTGASAVQIVPELVQL-AASVKVFQRTPGWVLPRVNFRHPAWARSTF
MAV_1795|M.avium_104 AVIGTGASAIQIIPELVKQ-AGFVKVFQRTPCWVLPRLDVATPPAVQALF
Mvan_1519|M.vanbaalenii_PYR-1 AVIGTGASAVQIVPELVKT-AAFVKVFQRTPGWVLPRLDLPMPAAVQALF
MLBr_00065|M.leprae_Br4923 VVIGSGATAVTLVPALANSGAKHVTMLQRSPTYIVSQP--AKDKIAARLN
.::*:**:.: ::* : . : : **:* :::.:
Mb3075c|M.bovis_AF2122/97 RTVPGLRALLRLGIYWAQEALAYGMTKRPNTLKIIEAYAKYNIRRSVKDR
Rv3049c|M.tuberculosis_H37Rv RTVPGLRALLRLGIYWAQEALAYGMTKRPNTLKIIEAYAKYNIRRSVKDR
MUL_1880|M.ulcerans_Agy99 ENVPGLRALARLGIYWAQEALAFGMTKRPNVLKVGEAYAKYNIRRSVKDR
TH_4080|M.thermoresistible__bu ENIPGLRLALRGGIYALQEALAVGMTKYPGMLRIGHLLGKWNINRSVKDP
MSMEG_2310|M.smegmatis_MC2_155 SYVPGTRALLRAAIYWIHEGVGFAMTKEPRLLKIGEWMGRYNINRSIKDP
Mflv_4608|M.gilvum_PYR-GCK RRLPRVQNVGRQAWFWGHELMAVGMVWDTPVTTGIQLLAKANLRRQVSDT
MAB_4476c|M.abscessus_ATCC_199 GSSPAVQNLARQAWYWTHEAAAVGMVWDTPVTTLIQWIAKANLRRQISDP
MMAR_3139|M.marinum_M KRMPATEQALRGAWFWAHEVMAVGMVWDTAATSVIQAAAKANLRRQVKDT
MAV_1795|M.avium_104 AKVPAAQELARQALYWGHEASATALVWDTPLTSLVARLGKAHLRAQVKDP
Mvan_1519|M.vanbaalenii_PYR-1 AKVPATQEIARQALYWGHEASAAALVWNTPMSGLVARLGKAHLRQQVKDP
MLBr_00065|M.leprae_Br4923 RWLPDKYAYTAVRWKNILRQSALYGACQKWPQRMRNILLGHVARRLPKGY
* . . . . ..
Mb3075c|M.bovis_AF2122/97 ELRRKLTPRYRIGCKRILNS--STYYPAVADPKTELITDRIDRITHDGIV
Rv3049c|M.tuberculosis_H37Rv ELRRKLTPRYRIGCKRILNS--STYYPAVADPKTELITDRIDRITHDGIV
MUL_1880|M.ulcerans_Agy99 ELRRKLTPHYRIGCKRILNS--SSYYPAVANPKTRLITDRIARITRNGIV
TH_4080|M.thermoresistible__bu QLRAKLTPDYRLGCKRILNS--STYYPAVADPRTEVITESIDRITTDGIV
MSMEG_2310|M.smegmatis_MC2_155 ELRRKLTPSYRAGCKRILNS--DTYYRGIADPKTQVITEGIARMTPTGIV
Mflv_4608|M.gilvum_PYR-GCK WLRRQLTPNFRAGCKRMLMS--SDYYPALQRDNCKLVSWPIATLSPNGIR
MAB_4476c|M.abscessus_ATCC_199 WVRRQLTPDFRAGCKRMLMT--SDYYAALEKDNCKLITWPIATLSPNGIR
MMAR_3139|M.marinum_M WLRRQLTPQFRPGCKRMLMT--NDYYPALQADNCKLVSWPIATLAPNGIR
MAV_1795|M.avium_104 WLRRQLTPDFRPGCKRMLVS--SDYYPALQRDNCKLIDWPIATLSPAGIR
Mvan_1519|M.vanbaalenii_PYR-1 WMRRQLTPDFTPGCKRMLIS--SDYYPALQRDNCKLIDWPIATLSPAGIR
MLBr_00065|M.leprae_Br4923 DVQKHFGPHYNPWEQRLCLVPDGDLFRAIRKGTVDMVTDAIDRFTSTGIR
:: :: * : :*: . : .: :: * :: **
Mb3075c|M.bovis_AF2122/97 TADGTGREVFREADVIVYATGFHVTDSYTYVQIKGRHGEDLVDRWNREGI
Rv3049c|M.tuberculosis_H37Rv TADGTGREVFREADVIVYATGFHVTDSYTYVQIKGRHGEDLVDRWNREGI
MUL_1880|M.ulcerans_Agy99 TADG----VEHQADVIVYATGFHVTDSYTYVQIKGQHGEDLVDRWNREGI
TH_4080|M.thermoresistible__bu TSDG----TERAVDVIVYATGFHVSDSYKYVQIKGLNGEDIVDRFNREGP
MSMEG_2310|M.smegmatis_MC2_155 TNDG----VEHPVDIVVYATGFHVTDSYTYVDIKGAGGEDLVDRWNREGI
Mflv_4608|M.gilvum_PYR-GCK TADG----IEHELDCIVFATGFDVCKAGTPFPIRGAHGRELSDEWS-QGA
MAB_4476c|M.abscessus_ATCC_199 TADG----IEHDVDCIVFATGFDVCKAGTPFPIAGLDGRKLADEWS-GGA
MMAR_3139|M.marinum_M TADG----IEHEVDCIVFATGFDVCKRGTPFPILGRDGRKLEDAWS-EGR
MAV_1795|M.avium_104 TSDG----IEHHLDCIVFATGYDVHLTGPPFPVTGLGGRSLAAEWA-GGA
Mvan_1519|M.vanbaalenii_PYR-1 TSDG----VEHHLDAIVFATGYDVHLSGPPFPVAGLNGRSLQQEWA-GHA
MLBr_00065|M.leprae_Br4923 LKSG----NELRADIIVTATGLNLQLFGGAVATVDGQPVDLAQTMS----
.* * :* *** .: . . .:
Mb3075c|M.bovis_AF2122/97 GAHRGITVANMPNLFFLLGPNTG---LGHNSVVFMIESQIHYVADAIAKC
Rv3049c|M.tuberculosis_H37Rv GAHRGITVANMPNLFFLLGPNTG---LGHNSVVFMIESQIHYVADAIAKC
MUL_1880|M.ulcerans_Agy99 GAHRGIAVADVPNLFFLLGPNTG---LGHNSVVFMIESQIHYVADAIAKC
TH_4080|M.thermoresistible__bu MAHRGTAVADLPNAYLLLGPNTG---LGHNSMVYMIESQINYVIEAIKAV
MSMEG_2310|M.smegmatis_MC2_155 QAHRGVAVAGMPNLFFLLGPNTA---LGHNSVVFMIEQQIRYVGQAIAAV
Mflv_4608|M.gilvum_PYR-GCK YAYRSVTVAGYPNLFFTFGPNSG---PGHNSALVYMEAEINYIVKAIGAI
MAB_4476c|M.abscessus_ATCC_199 YAYKSVNVSGYPNLFFTFGPNSG---PGHNSALVYMEAQIDYIVQAIGIV
MMAR_3139|M.marinum_M FAYKSVSVAGYPNLFFTFGPNSG---PGHNSALLYMEAAIDYIVKAIELL
MAV_1795|M.avium_104 QAYKSINVHGYPNLFVMTGPNSG---PGHNSLLVYIEGQLDYAVRGITTI
Mvan_1519|M.vanbaalenii_PYR-1 EAYKSASAHGYPNLFFMTGPNSG---PGHNSLLVYVEGQIAYAVDAITTI
MLBr_00065|M.leprae_Br4923 --YKGMMLSGLPNMIYTVGYTNASWTLKADLVSEFFCRLLNYMNANDFDT
::. . ** * ... : . : *
Mb3075c|M.bovis_AF2122/97 DR--MGVQALAPTREAQDRFNQELQRRLAGSVWNSGGCRSWYLDEHGKNT
Rv3049c|M.tuberculosis_H37Rv DR--MGVQALAPTREAQDRFNQELQRRLAGSVWNSGGCRSWYLDEHGKNT
MUL_1880|M.ulcerans_Agy99 DK--RGAQALAPTRAAQDRFNEELQRRLAGSVWNSGGCSSWYLDEHGRNT
TH_4080|M.thermoresistible__bu DK--AGAQALAPTREAQDRYNAELQARLGRSVWNTGGCRSWYLDEHGNNR
MSMEG_2310|M.smegmatis_MC2_155 DK--AGAAALAPTRRAQDAFNAELQRDLSGTVFNTGGCKSWYLDEHGVNR
Mflv_4608|M.gilvum_PYR-GCK I--ANDLSTLEVREDRQNNYHGEVQQRLGSTTWNSG-CKSWYLTNDGYNG
MAB_4476c|M.abscessus_ATCC_199 R--AKDLLTLDVRKDRQDRFNEDIQRRLGKTTWNSG-CQSWYLTEDGKNT
MMAR_3139|M.marinum_M RDQGNGVHTLDVKENSQNAYHSNIQRRLRRTTWNSG-CSSWYLTEDGYNG
MAV_1795|M.avium_104 LN--DDLRYLDVREEVQRRHNEAIQRRLTKTTWMSG-CRSWYLTKDGFNG
Mvan_1519|M.vanbaalenii_PYR-1 LN--RNLRYLDVRADVQQRHNAQIQRRLTKTTWMSG-CHSWYLTADGFNA
MLBr_00065|M.leprae_Br4923 VA--VEYPGSDVEERPFMDFTPGYVLRSLDALPKQGSRTPWRLNQNYLRD
. : * .* * . .
Mb3075c|M.bovis_AF2122/97 VLWCGYTWQYWLTTRSVNPAEYRFFGIGNG----------LSSDRATVAA
Rv3049c|M.tuberculosis_H37Rv VLWCGYTWQYWLTTRSVNPAEYRFFGIGNG----------LSSDRATVAA
MUL_1880|M.ulcerans_Agy99 VLWCGYTWQYWLATRSVKPEEYQFFKAG---------------AKSSVNG
TH_4080|M.thermoresistible__bu VLWGGFTWQYRLATRTLDPAEYRFWG------------------------
MSMEG_2310|M.smegmatis_MC2_155 TLWSGMTWQYWKALRRLDPSEFEFLDQR-------------VLARS----
Mflv_4608|M.gilvum_PYR-GCK TMYPGFATQFKRALSKVDMGDYVTTPTTARTEHHPTEHAQNGSNVAKVAQ
MAB_4476c|M.abscessus_ATCC_199 TMFPGFATQFARQLRTVELDDYSFT---------------SGAAAASAAK
MMAR_3139|M.marinum_M TMYPGFATQFTRELSRLDMRDYDITR--------------RDDADLKLTQ
MAV_1795|M.avium_104 SMYPGFATQYLRQMSDFRYQDYQAVAR---------------RARTPAAS
Mvan_1519|M.vanbaalenii_PYR-1 SMYPGFATQYRRQM-QFRAEDYTAVA------------------------
MLBr_00065|M.leprae_Br4923 IRLIRRGKVADEGLLFAKKTTPVTV-------------------------
Mb3075c|M.bovis_AF2122/97 AN---------------------------------------------
Rv3049c|M.tuberculosis_H37Rv AN---------------------------------------------
MUL_1880|M.ulcerans_Agy99 -----------------------------------------------
TH_4080|M.thermoresistible__bu -----------------------------------------------
MSMEG_2310|M.smegmatis_MC2_155 -----------------------------------------------
Mflv_4608|M.gilvum_PYR-GCK GRKAKVAQGRKSAGALPTRSDVKADILDVGVGKVPPKRAASRVRKDA
MAB_4476c|M.abscessus_ATCC_199 PP------------------------VDASIG---------------
MMAR_3139|M.marinum_M TR---------------------------------------------
MAV_1795|M.avium_104 SA---------------------------------------------
Mvan_1519|M.vanbaalenii_PYR-1 -----------------------------------------------
MLBr_00065|M.leprae_Br4923 -----------------------------------------------