For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DAAVVSQLSHVPAAARTLEQRGYDGCWTAEINHDPFLPLTLAAEHTERMELGTSIAVAFARNPMSVAQIG WDLQDYSEGRLLLGLGSQIKPHIEKRFSMPWGRPVARMREFVQALHEIWACWRDGTRLNFEGEFYTHTLM TPMFVPQQHAYADPKVFVAAVGDRMTEMCGEVADGLLAHAFSTQRYVREVTIPTLERGIARAGRTRADVV VSSPLFVVTGHDEQELAANAVATRKQIAFYASTPAYRGVLELHGWGDLQTELHRLSRAGEWDTMGALIDD TMLAEFAVVAPVDELVHTIRARCDGLIDRVLVGFPPSIDEATVVGLVSELQAEETS*
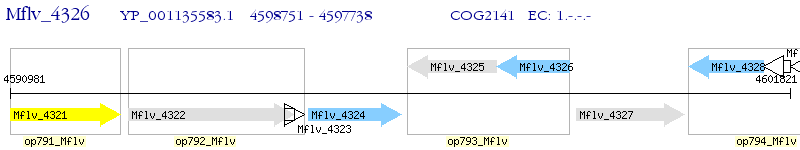
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4326 | - | - | 100% (337) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4500 | - | 2e-69 | 46.08% (319) | hypothetical protein Mflv_4500 |
| M. gilvum PYR-GCK | Mflv_5162 | - | 4e-42 | 31.21% (298) | luciferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1395 | - | 1e-120 | 63.55% (321) | oxidoreductase |
| M. tuberculosis H37Rv | Rv1360 | - | 1e-120 | 63.55% (321) | oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 9e-19 | 25.67% (300) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3914c | - | 3e-84 | 46.85% (333) | oxidoreductase |
| M. marinum M | MMAR_1390 | - | 5e-67 | 41.67% (324) | oxidoreductase |
| M. avium 104 | MAV_4719 | - | 1e-64 | 41.36% (324) | hypothetical protein MAV_4719 |
| M. smegmatis MC2 155 | MSMEG_2256 | - | 1e-135 | 68.37% (332) | hypothetical protein MSMEG_2256 |
| M. thermoresistible (build 8) | TH_1626 | - | 1e-139 | 69.07% (333) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_2486 | - | 4e-66 | 41.36% (324) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_2020 | - | 1e-163 | 84.04% (332) | putative oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1395|M.bovis_AF2122/97 ---MGGAR--RLKLDGSIPNQLARAADAAVALERNGFDGGWTAEAS-HDP
Rv1360|M.tuberculosis_H37Rv ---MGGAR--RLKLDGSIPNQLARAADAAVALERNGFDGGWTAEAS-HDP
Mflv_4326|M.gilvum_PYR-GCK -------------MDAAVVSQLSHVPAAARTLEQRGYDGCWTAEIN-HDP
Mvan_2020|M.vanbaalenii_PYR-1 ---MGAGG--GLKVDAAVVSQLSQVPAAARRLERRGYDGCWTAEIN-HDP
MSMEG_2256|M.smegmatis_MC2_155 ---MTDAQNPGVKVDRGISSRLSDVAQTARDLETAGCDGCWTGEIN-HDP
TH_1626|M.thermoresistible__bu ---MGGGAVSGLKIDRGIPSQLDRVPDVARTLERQGYDGCWTGEIN-HDP
MAB_3914c|M.abscessus_ATCC_199 -----------MLIDVTLTTGLNTMAAEAAEAEESGYAGIWTFEGA-HDP
MMAR_1390|M.marinum_M -----------MRVFTALYG-LADAAERARELRDAGATGVATFEGP-HDV
MUL_2486|M.ulcerans_Agy99 -----------MRVFTALYG-LADAAERARELRDAGASGVATFEGP-HDV
MAV_4719|M.avium_104 -----------MKVQLQVDGSPVRAAASAAEIAATGADGLFTFEGQ-HDV
MLBr_00348|M.leprae_Br4923 MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAWGSDA
: . . * . * * *
Mb1395|M.bovis_AF2122/97 FLPLLLAAEHTSRLELGTNIAVAFARNPMIVANVGWDLQTYSKGRLILGL
Rv1360|M.tuberculosis_H37Rv FLPLLLAAEHTSRLELGTNIAVAFARNPMIVANVGWDLQTYSKGRLILGL
Mflv_4326|M.gilvum_PYR-GCK FLPLTLAAEHTERMELGTSIAVAFARNPMSVAQIGWDLQDYSEGRLLLGL
Mvan_2020|M.vanbaalenii_PYR-1 FLPLTLAAEHTHSIELGTSIAVAFARNPMTVAQIGWDLQDYSGGRFILGL
MSMEG_2256|M.smegmatis_MC2_155 FLPLALAAEHTRRIQIGTSIAVAFARNPMTVAHLGWDLQAYSGGRFILGL
TH_1626|M.thermoresistible__bu FLPLLLAAEHTTRLEIGTSIAVAFARNPMTVANIGWDLQAYSRGRFVLGL
MAB_3914c|M.abscessus_ATCC_199 FLPLLLAAEHTKNVMLGTSIAVAFARNPMLLATIGWDLQAYSGGRFILGL
MMAR_1390|M.marinum_M FAPLTLAATVGG-LDLMTNVAIAFPRNPIHLAHQAYDHQLLSGGRFILGL
MUL_2486|M.ulcerans_Agy99 FAPLTLAATVGG-LDLMTNVAIAFPRNPIHLAHQAYDHQLLSGGRFILGL
MAV_4719|M.avium_104 FFPLLIAAEATG-CELMTNVAIAPPRSPMHLAYAAYDLQLYSGGRFRLGL
MLBr_00348|M.leprae_Br4923 YTPLAWWGSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
: ** . : *.:. .*.* * . : * ** ***
Mb1395|M.bovis_AF2122/97 GTQIRPHIEKRFSMPWGHPARRMREFVAALRAIWLAWQ----DGTKLCFE
Rv1360|M.tuberculosis_H37Rv GTQIRPHIEKRFSMPWGHPARRMREFVAALRAIWLAWQ----DGTKLCFE
Mflv_4326|M.gilvum_PYR-GCK GSQIKPHIEKRFSMPWGRPVARMREFVQALHEIWACWR----DGTRLNFE
Mvan_2020|M.vanbaalenii_PYR-1 GSQIKPHIEKRFSMPWSKPVGRMREFVLALRAIWMSWS----DGSRLEFD
MSMEG_2256|M.smegmatis_MC2_155 GTQIRPHIEKRFSMPWSHPVRRMREYIAALQAIWTCWR----DGTSLRFE
TH_1626|M.thermoresistible__bu GTQIRPHVERRFSMPWSHPVRRMREFVTALREIWSCWR----DGTRLSFH
MAB_3914c|M.abscessus_ATCC_199 GTQIKPHITRRFDMPWSSPAARMRDMVLAVRAIWQSWQ----ERGALDYR
MMAR_1390|M.marinum_M GTQVRAQIENRFGAAFDHPVARMTELIAALRAIFAAWN----CGDRLNFR
MUL_2486|M.ulcerans_Agy99 GTQVRAQIENRFGAAFDHPVARMTELIAALRAIFAAWN----CGDRLNFR
MAV_4719|M.avium_104 GSQIKVHVEKRYGSKWEPPAARMAEAISAIKAIFAAWE----GRARLDFR
MLBr_00348|M.leprae_Br4923 GVSGPQVVEGWYGQPFPKPLSRTREYIDIVRQVWAREAPVVSDGQHYPLP
* . : :. : * * : : :: ::
Mb1395|M.bovis_AF2122/97 GEFYTHKIMTPMFTPEP---QPYPVPRVFIAAVGEAMTEMCGEVADGHLG
Rv1360|M.tuberculosis_H37Rv GEFYTHKIMTPMFTPEP---QPYPVPRVFIAAVGEAMTEMCGEVADGHLG
Mflv_4326|M.gilvum_PYR-GCK GEFYTHTLMTPMFVPQQ---HAYADPKVFVAAVGDRMTEMCGEVADGLLA
Mvan_2020|M.vanbaalenii_PYR-1 GEFYTHKLMTPMFVPPR---HPYGDPRVFVAAVGDRMTEMCGEVADGLLA
MSMEG_2256|M.smegmatis_MC2_155 GEFYTHKIMTPMFTPEP---SPAPFPKVFVAAVGERMTEMCGEAADGLLA
TH_1626|M.thermoresistible__bu GDFYSHTLMTPMFVPEP---QPHGDPRIFLAAVGPEMTRMCGEVADGLLA
MAB_3914c|M.abscessus_ATCC_199 GDFYQNTLMTPMFMPDPAEVSAFGPPPIWLAGVGTGMTEVAGEVADGFMS
MMAR_1390|M.marinum_M GQFYRHTLMTPTFNPGP---IPFGPPPIYVGALGPRLTRAVAQHADGLLV
MUL_2486|M.ulcerans_Agy99 GQFYRHTLMTPTSNPGP---IPFGPPPIYVGALGPRLTRAVAQHADGLLV
MAV_4719|M.avium_104 GEFYTHTIMAPTFDPGP---NPYGPPPVLLGALGPMMTRTAAEVADGLLV
MLBr_00348|M.leprae_Br4923 LTGAGATGLGKALKPIT--HPLRADIPIMLGAEGPKNVALAAEICDGWLP
. : * : :.. * . .: .** :
Mb1395|M.bovis_AF2122/97 HPMVSKRYLTEVSVPALLRGLARSG--RDRSAFEVSCEVMVATGADDAEL
Rv1360|M.tuberculosis_H37Rv HPMVSKRYLTEVSVPALLRGLARSG--RDRSAFEVSCEVMVATGADDAEL
Mflv_4326|M.gilvum_PYR-GCK HAFSTQRYVREVTIPTLERGIARAG--RTRADVVVSSPLFVVTGHDEQEL
Mvan_2020|M.vanbaalenii_PYR-1 HAFSTQRYVREVTIPTLTRGIERAG--RTRADIEVASPLFIVTGLDEQQM
MSMEG_2256|M.smegmatis_MC2_155 HAFTTRRYLDEVTLPALQRGMDRTG--RSRNDFEVSCPVFLVTGSDEHQM
TH_1626|M.thermoresistible__bu HAFTTRRYLDEVTLPALLDGMNRAG--RRRGDFEVSCPVFVVTGTTETEL
MAB_3914c|M.abscessus_ATCC_199 HPFCTPDYLAQVTRPALGRGALKSG--KALADIQIHHSPMIIIGRDDAEL
MMAR_1390|M.marinum_M MPFGSKRFLHANTMPAVHEGLRAGG--RQAGEFTIIPEIIVSAGDTEADR
MUL_2486|M.ulcerans_Agy99 MPFGSKRFLHANTMPAVHEGLRAGG--RQAGDFTIIPEIIVSAGDTEADR
MAV_4719|M.avium_104 MPFNSARHFAERTLPAIGEGLRRSG--RTLTEFPIIAQAMVAVGRDEADL
MLBr_00348|M.leprae_Br4923 IFFSPR--MADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDR---
: . . : * * : . : :
Mb1395|M.bovis_AF2122/97 AAACTATRKQIAFYGS------TPAYRKVLEQHGWGDLHPELHRLSKLGE
Rv1360|M.tuberculosis_H37Rv AAACTATRKQIAFYGS------TPAYRKVLEQHGWGDLHPELHRLSKLGE
Mflv_4326|M.gilvum_PYR-GCK AANAVATRKQIAFYAS------TPAYRGVLELHGWGDLQTELHRLSRAGE
Mvan_2020|M.vanbaalenii_PYR-1 AAAAVATRKQIAFYAS------TPAYRSVLELHGWGDLQTELHSLSREGD
MSMEG_2256|M.smegmatis_MC2_155 AEAAVATRKQLAFYGS------TPAYRKVLELHGWGDLHTELHRLSLAGE
TH_1626|M.thermoresistible__bu RANAVATRRQLAFYAS------TPAYRSVLDLHGWGDLHTELHALSRRGE
MAB_3914c|M.abscessus_ATCC_199 ASARAAVRKQLAFYGS------TPAYWPILELHGRAEMGPELKERSKQGE
MMAR_1390|M.marinum_M EQAAAGTRRLLAFYGS------TPAYRPVLAAHGWEDLQPELNALSKQGR
MUL_2486|M.ulcerans_Agy99 EQAAAGTRRLLAFYGS------TPAYRPVLAAHGWEGLQPELNALSKQGR
MAV_4719|M.avium_104 ATAIDGVASLIAFYGS------TPAYLPVLQVEGWDGIQPELNALSKQGR
MLBr_00348|M.leprae_Br4923 AAAFAGIKPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQ
. :*:* . * : : * : *: .
Mb1395|M.bovis_AF2122/97 WEAMGGLIDDEMLGAFAVVGP-VDTIAGALRNRCEGVVDRVLPIFMAAS-
Rv1360|M.tuberculosis_H37Rv WEAMGGLIDDEMLGAFAVVGP-VDTIAGALRNRCEGVVDRVLPIFMAAS-
Mflv_4326|M.gilvum_PYR-GCK WDTMGALIDDTMLAEFAVVAP-VDELVHTIRARCDGLIDRVLVGFPPSID
Mvan_2020|M.vanbaalenii_PYR-1 WDTMGSLIDDGMLAEFAVVAL-VDDVVDKIRARCDGLIDRVLVGFPPSID
MSMEG_2256|M.smegmatis_MC2_155 WDTMGTLIDDDVLDAFAVVAP-LDELADALRRRCEGAIDRVLPGFPAAVP
TH_1626|M.thermoresistible__bu WDAMGERIDDDVLAAFAVVAP-LDELAGRLRERCAGVIDRVLPAFPAGVP
MAB_3914c|M.abscessus_ATCC_199 WEAMAALVDDDFVDTAAVTVTDPGQGARELQERYGDLVHRLGFNTPYRPD
MMAR_1390|M.marinum_M WQEMGALIDDTMMHTLAACGT-PADVAAHIRDRVDGISDTVCLYQAAPIG
MUL_2486|M.ulcerans_Agy99 WQEMGALIDDTMMHTLAACGT-PADVAARIRDRVDGISDTVCLYQAAPIG
MAV_4719|M.avium_104 FAQMRGLITDELVARIGIVGT-PEQCAEQIAARFGEHADEVCCYFPGYTP
MLBr_00348|M.leprae_Br4923 KDKAAEIIPNELVDSTMIVGD-VDYVCKQIAAWSAAGVTMMMVSASSVEQ
: : .: : :
Mb1395|M.bovis_AF2122/97 -QECINAALQDFRR-----------------
Rv1360|M.tuberculosis_H37Rv -QECINAALQDFRR-----------------
Mflv_4326|M.gilvum_PYR-GCK -EATVVGLVSELQAEETS-------------
Mvan_2020|M.vanbaalenii_PYR-1 -EATVVDLVTDLRTGA---------------
MSMEG_2256|M.smegmatis_MC2_155 -PATVNAVLDEFRDRP-----PVTRL-----
TH_1626|M.thermoresistible__bu -ETAVTAVLHELRAAPSPTTSPVSTERSRHP
MAB_3914c|M.abscessus_ATCC_199 -RALLPELLAAFPTR----------------
MMAR_1390|M.marinum_M -LRTLAAIIDELR------------------
MUL_2486|M.ulcerans_Agy99 -LRTLAAIIDELR------------------
MAV_4719|M.avium_104 RPADVADLIGALHRVPALP------------
MLBr_00348|M.leprae_Br4923 -VRDLAGLF----------------------
: .