For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVFTALYGLADAAERARELRDAGASGVATFEGPHDVFAPLTLAATVGGLDLMTNVAIAFPRNPIHLAHQA YDHQLLSGGRFILGLGTQVRAQIENRFGAAFDHPVARMTELIAALRAIFAAWNCGDRLNFRGQFYRHTLM TPTSNPGPIPFGPPPIYVGALGPRLTRAVAQHADGLLVMPFGSKRFLHANTMPAVHEGLRAGGRQAGDFT IIPEIIVSAGDTEADREQAAAGTRRLLAFYGSTPAYRPVLAAHGWEGLQPELNALSKQGRWQEMGALIDD TMMHTLAACGTPADVAARIRDRVDGISDTVCLYQAAPIGLRTLAAIIDELR*
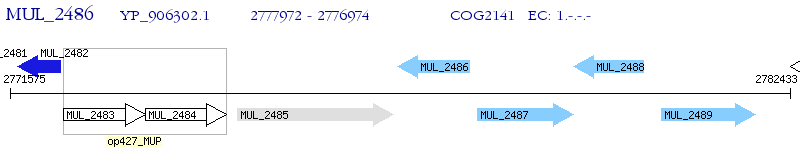
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2486 | - | - | 100% (332) | oxidoreductase |
| M. ulcerans Agy99 | MUL_2504 | - | 5e-44 | 32.70% (315) | hypothetical protein MUL_2504 |
| M. ulcerans Agy99 | MUL_4782 | - | 9e-19 | 28.34% (307) | coenzyme F420-dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1395 | - | 7e-70 | 45.54% (303) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4500 | - | 1e-134 | 71.65% (328) | hypothetical protein Mflv_4500 |
| M. tuberculosis H37Rv | Rv1360 | - | 7e-70 | 45.54% (303) | oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 3e-10 | 25.37% (339) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3914c | - | 1e-60 | 41.78% (304) | oxidoreductase |
| M. marinum M | MMAR_1390 | - | 0.0 | 98.49% (332) | oxidoreductase |
| M. avium 104 | MAV_4719 | - | 3e-92 | 54.64% (302) | hypothetical protein MAV_4719 |
| M. smegmatis MC2 155 | MSMEG_2037 | - | 1e-140 | 72.29% (332) | hypothetical protein MSMEG_2037 |
| M. thermoresistible (build 8) | TH_3181 | - | 1e-134 | 71.30% (331) | PUTATIVE conserved hypothetical protein |
| M. vanbaalenii PYR-1 | Mvan_1819 | - | 1e-135 | 71.90% (331) | hypothetical protein Mvan_1819 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1395|M.bovis_AF2122/97 -----MGGARRLKLDGSIPNQLARAADAAVALERNGFDGGWTAEAS-HDP
Rv1360|M.tuberculosis_H37Rv -----MGGARRLKLDGSIPNQLARAADAAVALERNGFDGGWTAEAS-HDP
MAB_3914c|M.abscessus_ATCC_199 -----------MLIDVTLTTGLNTMAAEAAEAEESGYAGIWTFEGA-HDP
MUL_2486|M.ulcerans_Agy99 -----------MRVFTALYG-LADAAERARELRDAGASGVATFEGP-HDV
MMAR_1390|M.marinum_M -----------MRVFTALYG-LADAAERARELRDAGATGVATFEGP-HDV
Mflv_4500|M.gilvum_PYR-GCK --------------MTALFG-PIDAVERARALREAGAAGVFTFEGP-HDV
Mvan_1819|M.vanbaalenii_PYR-1 -----------MKIMTALFG-PTDAIERAALLREAGASGVFTFEGP-FDV
MSMEG_2037|M.smegmatis_MC2_155 -----------MKIMTALFG-PTDAIERTHALVEAGATGVFTFEGP-HDV
TH_3181|M.thermoresistible__bu -----------VKVMTALFG-PIDAAEQARTLREAGVAGVFTFEGP-HDV
MAV_4719|M.avium_104 -----------MKVQLQVDGSPVRAAASAAEIAATGADGLFTFEGQ-HDV
MLBr_00348|M.leprae_Br4923 MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAWGSDA
. * . * *. *
Mb1395|M.bovis_AF2122/97 FLPLLLAAEHTSRLELGTNIAVAFARNPMIVANVGWDLQTYSKGRLILGL
Rv1360|M.tuberculosis_H37Rv FLPLLLAAEHTSRLELGTNIAVAFARNPMIVANVGWDLQTYSKGRLILGL
MAB_3914c|M.abscessus_ATCC_199 FLPLLLAAEHTKNVMLGTSIAVAFARNPMLLATIGWDLQAYSGGRFILGL
MUL_2486|M.ulcerans_Agy99 FAPLTLAATVGG-LDLMTNVAIAFPRNPIHLAHQAYDHQLLSGGRFILGL
MMAR_1390|M.marinum_M FAPLTLAATVGG-LDLMTNVAIAFPRNPIHLAHQAYDHQLLSGGRFILGL
Mflv_4500|M.gilvum_PYR-GCK FTPLTLASTVGG-LDLMTNVAIAFPRNPIHLAHQAVDHQLLSGGRFTLGL
Mvan_1819|M.vanbaalenii_PYR-1 FTPLVLASTVKG-LDSMSNVAIAFPRNPIQLAHQANDLQLLTEGRFILGL
MSMEG_2037|M.smegmatis_MC2_155 FAPLTLASTVGG-VDLMTNVAIAFPRNPIHLAHQAIDHQLLSGGRFTLGL
TH_3181|M.thermoresistible__bu FTPLVRAATEPG-LELLTNVAIAFPRNPIHLAHQAYDLQLLSGGRFTLGL
MAV_4719|M.avium_104 FFPLLIAAEATG-CELMTNVAIAPPRSPMHLAYAAYDLQLYSGGRFRLGL
MLBr_00348|M.leprae_Br4923 YTPLAWWGSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
: ** . :.:. .*.* * . : : ** ***
Mb1395|M.bovis_AF2122/97 GTQIRPHIEKRFSMPWGHPARRMREFVAALRAIWLAWQDGTKLCFEGEFY
Rv1360|M.tuberculosis_H37Rv GTQIRPHIEKRFSMPWGHPARRMREFVAALRAIWLAWQDGTKLCFEGEFY
MAB_3914c|M.abscessus_ATCC_199 GTQIKPHITRRFDMPWSSPAARMRDMVLAVRAIWQSWQERGALDYRGDFY
MUL_2486|M.ulcerans_Agy99 GTQVRAQIENRFGAAFDHPVARMTELIAALRAIFAAWNCGDRLNFRGQFY
MMAR_1390|M.marinum_M GTQVRAQIENRFGAAFDHPVARMTELIAALRAIFAAWNCGDRLNFRGQFY
Mflv_4500|M.gilvum_PYR-GCK GTQIRTQIEKRYGTDFDRPVARMRDLVGALRAIFHTWSTGDRLAYRGEFY
Mvan_1819|M.vanbaalenii_PYR-1 GTQVRAQIEKRYGAAFDRPVERMKEMVGALRAIFAAWNEGERLDFRGEFY
MSMEG_2037|M.smegmatis_MC2_155 GTQIRTQIEKRFGADFERPVGRMRELIGALRAIFDAWQTGERLDFRGEFY
TH_3181|M.thermoresistible__bu GTQIRTQIEKRFGAEFDRPVARMKELVAALRQIFAAWQHGERLDFRGEFY
MAV_4719|M.avium_104 GSQIKVHVEKRYGSKWEPPAARMAEAISAIKAIFAAWEGRARLDFRGEFY
MLBr_00348|M.leprae_Br4923 GVSGPQVVEGWYGQPFPKPLSRTREYIDIVR---QVWAREAPVVSDGQHY
* . : :. : * * : : :: * : *:.*
Mb1395|M.bovis_AF2122/97 THKIMTPMFTPEP---QPYPVP-----RVFIAAVGEAMTEMCGEVADGHL
Rv1360|M.tuberculosis_H37Rv THKIMTPMFTPEP---QPYPVP-----RVFIAAVGEAMTEMCGEVADGHL
MAB_3914c|M.abscessus_ATCC_199 QNTLMTPMFMPDPAEVSAFGPP-----PIWLAGVGTGMTEVAGEVADGFM
MUL_2486|M.ulcerans_Agy99 RHTLMTPTSNPGP---IPFGPP-----PIYVGALGPRLTRAVAQHADGLL
MMAR_1390|M.marinum_M RHTLMTPTFNPGP---IPFGPP-----PIYVGALGPRLTRAVAQHADGLL
Mflv_4500|M.gilvum_PYR-GCK RHTLMTPTFTPR----APFDPI-----PIYVGALGPRLTRMTAEVADGLL
Mvan_1819|M.vanbaalenii_PYR-1 RHTLMTPTFVPGP---NPFGPP-----PIYLGALGPRLTRATAEVADGLL
MSMEG_2037|M.smegmatis_MC2_155 RHTLMTPTFSPGP---SPYGPP-----PIYVGALGPQLTRATAELADGLL
TH_3181|M.thermoresistible__bu RHTLMTPTFNPGP---NPYGPP-----PIQLGALGPQLTRAAAEVADGLL
MAV_4719|M.avium_104 THTIMAPTFDPGP---NPYGPP-----PVLLGALGPMMTRTAAEVADGLL
MLBr_00348|M.leprae_Br4923 PLPLTGAGATGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWL
: . . : :.. * . .: .** :
Mb1395|M.bovis_AF2122/97 GHPMVSKRYLTEVSVPALLRGLARSG--RDRSAFEVSCEVMVATGADDAE
Rv1360|M.tuberculosis_H37Rv GHPMVSKRYLTEVSVPALLRGLARSG--RDRSAFEVSCEVMVATGADDAE
MAB_3914c|M.abscessus_ATCC_199 SHPFCTPDYLAQVTRPALGRGALKSG--KALADIQIHHSPMIIIGRDDAE
MUL_2486|M.ulcerans_Agy99 VMPFGSKRFLHANTMPAVHEGLRAGG--RQAGDFTIIPEIIVSAGDTEAD
MMAR_1390|M.marinum_M VMPFGSKRFLHANTMPAVHEGLRAGG--RQAGEFTIIPEIIVSAGDTEAD
Mflv_4500|M.gilvum_PYR-GCK VMPFGSRRFLTEVTMPNVAEGLAAAG--RTRADLAVVPEIIVSVAD-QDP
Mvan_1819|M.vanbaalenii_PYR-1 VMPFGSKRFLHGTTMPAVRDGLAAAG--RSEDDFEVVPEIIVSVATGSDD
MSMEG_2037|M.smegmatis_MC2_155 VMPFGSKRFLHEVTVPAVRAGLNAAG--RSADDFAVVPEIIVSVGE-DDR
TH_3181|M.thermoresistible__bu VMPFGTTRYLRERTMAAVDAGLAAAG--RRREDFAVVPEIIVSPGD----
MAV_4719|M.avium_104 VMPFNSARHFAERTLPAIGEGLRRSG--RTLTEFPIIAQAMVAVGRDEAD
MLBr_00348|M.leprae_Br4923 PIFFSPR--MADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAA
: . : : * * : : : . :
Mb1395|M.bovis_AF2122/97 LAAACTATRKQIAFYG------STPAYRKVLEQHGWGDLHPELHRLSKLG
Rv1360|M.tuberculosis_H37Rv LAAACTATRKQIAFYG------STPAYRKVLEQHGWGDLHPELHRLSKLG
MAB_3914c|M.abscessus_ATCC_199 LASARAAVRKQLAFYG------STPAYWPILELHGRAEMGPELKERSKQG
MUL_2486|M.ulcerans_Agy99 REQAAAGTRRLLAFYG------STPAYRPVLAAHGWEGLQPELNALSKQG
MMAR_1390|M.marinum_M REQAAAGTRRLLAFYG------STPAYRPVLAAHGWEDLQPELNALSKQG
Mflv_4500|M.gilvum_PYR-GCK ---DHMSARRLLAFYG------STPAYRPVLEVHGWGDLQPELNALSKQG
Mvan_1819|M.vanbaalenii_PYR-1 ---DHASTRMLLAFYG------STPAYRPVLDAHGWGDLQPELNAMSKQG
MSMEG_2037|M.smegmatis_MC2_155 ---DHDATRRLLAFYG------STPAYRPVLEAHGWGDLQPELNAMSKQG
TH_3181|M.thermoresistible__bu ---DHTATRRLLAFYG------STPAYRPVLDLHGWGDLQPELNALSKQG
MAV_4719|M.avium_104 LATAIDGVASLIAFYG------STPAYLPVLQVEGWDGIQPELNALSKQG
MLBr_00348|M.leprae_Br4923 ---AFAGIKPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSN
. :*:* .* : : * : *: : .
Mb1395|M.bovis_AF2122/97 EWEAMGGLIDDEMLG-AFAVVGPVDTIAGALRNRCEGVVDRV-LPIFMAA
Rv1360|M.tuberculosis_H37Rv EWEAMGGLIDDEMLG-AFAVVGPVDTIAGALRNRCEGVVDRV-LPIFMAA
MAB_3914c|M.abscessus_ATCC_199 EWEAMAALVDDDFVDTAAVTVTDPGQGARELQERYGDLVHRLGFNTPYRP
MUL_2486|M.ulcerans_Agy99 RWQEMGALIDDTMMH-TLAACGTPADVAARIRDRVDGISDTVCLYQAAPI
MMAR_1390|M.marinum_M RWQEMGALIDDTMMH-TLAACGTPADVAAHIRDRVDGISDTVCLYQAAPI
Mflv_4500|M.gilvum_PYR-GCK RWEDMGRLIDDDVLH-TIAACGTPADVAKHIRDRVDGVSDRICLYQPGPI
Mvan_1819|M.vanbaalenii_PYR-1 RWQEMATLIDDEILH-TIAACGSPAEVADHIRDRVDGVSDRLCLYQPGPI
MSMEG_2037|M.smegmatis_MC2_155 RWQEMSALIDDDVLH-TIAACGSPAEIAAHIADRVAGVGDRVCLYQPTPI
TH_3181|M.thermoresistible__bu RWQEMGALIDDDILH-TVAACGGPAEIAAHIRERVAGISDRVCLYAAGPI
MAV_4719|M.avium_104 RFAQMRGLITDELVA-RIGIVGTPEQCAEQIAARFGEHADEVCCYFPGYT
MLBr_00348|M.leprae_Br4923 QKDKAAEIIPNELVD-STMIVGDVDYVCKQIAAWSAAGVTMMMVSASSVE
. :: : .: . : :
Mb1395|M.bovis_AF2122/97 S-QECINAALQDFRR-----
Rv1360|M.tuberculosis_H37Rv S-QECINAALQDFRR-----
MAB_3914c|M.abscessus_ATCC_199 D-RALLPELLAAFPTR----
MUL_2486|M.ulcerans_Agy99 G-LRTLAAIIDELR------
MMAR_1390|M.marinum_M G-LRTLAAIIDELR------
Mflv_4500|M.gilvum_PYR-GCK A-VNSLAEIVDALRG-----
Mvan_1819|M.vanbaalenii_PYR-1 A-VDALAEIVDALS------
MSMEG_2037|M.smegmatis_MC2_155 S-LDSLAQIVDGLRT-----
TH_3181|M.thermoresistible__bu P-VERLAAIVHELAD-----
MAV_4719|M.avium_104 PRPADVADLIGALHRVPALP
MLBr_00348|M.leprae_Br4923 Q-VRDLAGLF----------
: .