For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVLGDYRPGGSWLHRLPAGVKLIGLGLVIIAMTLAVDSPLRLGVATVVMVSVVASARLSPVALLAQLRPV LWVVGFIFVLQVALTGWRRALVVCGVLLLAVALAAIVTTTTRTADMLDAATRAMTPLARFGFPVRQVAIA LALTVRSIPLLVEIIRQMEEARRARGLRISPRIVFVPVIIGALRAADDFSEALIARGID*
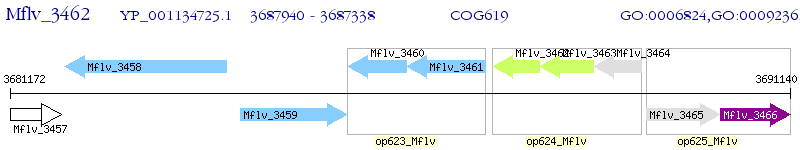
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3462 | - | - | 100% (200) | cobalt transport protein |
| M. gilvum PYR-GCK | Mflv_2731 | - | 1e-10 | 25.79% (221) | cobalt transport protein |
| M. gilvum PYR-GCK | Mflv_4056 | - | 8e-08 | 28.95% (190) | cobalt ABC transporter, inner membrane subunit CbiQ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2352c | - | 1e-13 | 29.15% (223) | hypothetical protein Mb2352c |
| M. tuberculosis H37Rv | Rv2325c | - | 1e-13 | 29.15% (223) | hypothetical protein Rv2325c |
| M. leprae Br4923 | MLBr_00849 | - | 8e-11 | 26.03% (219) | hypothetical protein MLBr_00849 |
| M. abscessus ATCC 19977 | MAB_1714 | - | 5e-12 | 28.96% (221) | hypothetical protein MAB_1714 |
| M. marinum M | MMAR_3625 | - | 3e-09 | 24.31% (218) | hypothetical protein MMAR_3625 |
| M. avium 104 | MAV_3956 | - | 4e-12 | 29.33% (225) | cobalt transport protein, putative |
| M. smegmatis MC2 155 | MSMEG_4469 | - | 7e-13 | 26.94% (219) | cobalt transport protein |
| M. thermoresistible (build 8) | TH_3631 | - | 7e-62 | 60.00% (200) | PUTATIVE cobalt transport protein |
| M. ulcerans Agy99 | MUL_1240 | - | 2e-09 | 24.31% (218) | hypothetical protein MUL_1240 |
| M. vanbaalenii PYR-1 | Mvan_3226 | - | 1e-80 | 75.00% (200) | cobalt transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2352c|M.bovis_AF2122/97 --------------------------------------------------
Rv2325c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3625|M.marinum_M --------------------------------------------------
MUL_1240|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00849|M.leprae_Br4923 --------------------------------------------------
MAV_3956|M.avium_104 MAGLTVPTTGTCLLDGRPAAEQVGAVALQFQAARLQLMRSRVDLEVASAA
MSMEG_4469|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1714|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3462|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3226|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3631|M.thermoresistible__bu --------------------------------------------------
Mb2352c|M.bovis_AF2122/97 --------------------------------------------------
Rv2325c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3625|M.marinum_M --------------------------------------------------
MUL_1240|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00849|M.leprae_Br4923 --------------------------------------------------
MAV_3956|M.avium_104 GFSPDDHDRVSAALATVGLDAGLAERRIDQLSGGQMRRVVLAGLLARSPR
MSMEG_4469|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1714|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3462|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3226|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3631|M.thermoresistible__bu --------------------------------------------------
Mb2352c|M.bovis_AF2122/97 --------------------------------------------------
Rv2325c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3625|M.marinum_M --------------------------------------------------
MUL_1240|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00849|M.leprae_Br4923 --------------------------------------------------
MAV_3956|M.avium_104 VLILDEPLAGLDAASQRGLVELLAERRRETGLTVVVISHDFAGLEQLCPR
MSMEG_4469|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1714|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3462|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3226|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3631|M.thermoresistible__bu --------------------------------------------------
Mb2352c|M.bovis_AF2122/97 --------------MTTTS----APARNGTRRPSRPIVLLIPVPGSSVIH
Rv2325c|M.tuberculosis_H37Rv --------------MTTTS----APARNGTRRPSRPIVLLIPVPGSSVIH
MMAR_3625|M.marinum_M --------------MTVSSPSASATPERGSRRAARPVVLLVPVPRGSAIH
MUL_1240|M.ulcerans_Agy99 --------------MTVSSPSASATPERGSRRATRPVVLLVPAPRGSAIH
MLBr_00849|M.leprae_Br4923 --------------MTSLS----MSEHHVPRRRSRPVVLLVPVPGTSTIH
MAV_3956|M.avium_104 ILHLRDGSLDANPVAAQPVPVPVAPPTKRPAARRRPVVLLRPVPGSSPIH
MSMEG_4469|M.smegmatis_MC2_155 --------------------------MSAPGGQRKPVVLLRPVPGDSVIH
MAB_1714|M.abscessus_ATCC_1997 --------------------------MTKTRAPRRPVVLLRPVPGTSTIH
Mflv_3462|M.gilvum_PYR-GCK -----------------------------------MTVLGDYRPGGSWLH
Mvan_3226|M.vanbaalenii_PYR-1 -----------------------------------MTMLGDYRAGTSWLH
TH_3631|M.thermoresistible__bu -----------------------------------MSTFGQYQPGSSPLH
: . * :*
Mb2352c|M.bovis_AF2122/97 DLWAGTKLLVVFGISVLLTFYPG-WVTIGMMAALVLAAARIAHIPRGALP
Rv2325c|M.tuberculosis_H37Rv DLWAGTKLLVVFGISVLLTFYPG-WVTIGMMAALVLAAARIAHIPRGALP
MMAR_3625|M.marinum_M DLWAGTKLLVVFAISVLLTFYPG-WVTIGLMAALVLVAAWIAHIPRGAVP
MUL_1240|M.ulcerans_Agy99 DLWAGTKLLVVFAISVLLTFYPG-WVTIGLMAALVLVAAWIAHIPRGAVP
MLBr_00849|M.leprae_Br4923 DLWAGTKLLVVFGVEVLLTFYPG-WVTIGLVAALVCGAAWIARIPRDVLP
MAV_3956|M.avium_104 ELWAGTKLLVVFAMSLLLTVFPG-WVAIGLATALAAAGLRLAHIPRGVLP
MSMEG_4469|M.smegmatis_MC2_155 QLWAGTKLLSVAVIGIMLTFYPG-WVPIGFTAVLVLVVARLARIPRGVVP
MAB_1714|M.abscessus_ATCC_1997 ELWAGTKLIAVFLISVLLTFFPT-WTAIGAVAALVILTAWLAHIPRGAVP
Mflv_3462|M.gilvum_PYR-GCK RLPAGVKLIGLGLVIIAMTLAVDSPLRLGVATVVMVSVVASARLSPVALL
Mvan_3226|M.vanbaalenii_PYR-1 RMPAGAKLFGLGVLIVAMTVWVDSPVRLAVVAGIVLAAVLSARLNVRALI
TH_3631|M.thermoresistible__bu RLPAGVKLVALAVSIALMAVWVQTPADLAVAALGVAALVAAAGLRPRALT
: **.**. : ::. :. : * : .:
Mb2352c|M.bovis_AF2122/97 SVPRWLWIVLAIGFLTAALAGGTPVVAVGGVQLGLGGALHFLRITALSVV
Rv2325c|M.tuberculosis_H37Rv SVPRWLWIVLAIGFLTAALAGGTPVVAVGGVQLGLGGALHFLRITALSVV
MMAR_3625|M.marinum_M SMPRWLFVVLAFGGLTAALAGGDPMVSVGGVELGLGGALNFLRITALSIV
MUL_1240|M.ulcerans_Agy99 SMPRWLFVVLAFGGLTAALAGGDPMVSVGGVELGLGGALNFLRITALSIV
MLBr_00849|M.leprae_Br4923 SMPRWLWIVVVLGGFTAALAGGSPVVSMGGVDIGLGGVLNFLRITAWSVV
MAV_3956|M.avium_104 SVPRWLWIFLGVVGVTAALAGGAPTIHLGTASLGLGGLLDFLRATALTVV
MSMEG_4469|M.smegmatis_MC2_155 TIPWWMWLLVAFGALTATFAGGSPEFDVGSMTIGLGGLLNFLRITALSIV
MAB_1714|M.abscessus_ATCC_1997 TIPIWMVALVIGGSLTVFFSGGPPFWHIGGLTIGFHGVLAVARTTAVAAV
Mflv_3462|M.gilvum_PYR-GCK AQLRPVLWVVGF---------------IFVLQVALTGWRRALVVCGVLLL
Mvan_3226|M.vanbaalenii_PYR-1 LQLRQVLWVVGF---------------IFVLQVVLTDLHRALVVCGVLLL
TH_3631|M.thermoresistible__bu AQLRPVLWVVAV---------------IFGFQLLFTDWRRALVVCGILLL
: .: : : : . . :
Mb2352c|M.bovis_AF2122/97 LLALGAMVSWTTNVAEISPAVATLGRPFRVLRIPVDEWAVALALALRAFP
Rv2325c|M.tuberculosis_H37Rv LLALGAMVSWTTNVAEISPAVATLGRPFRVLRIPVDEWAVALALALRAFP
MMAR_3625|M.marinum_M LLALGAMVSWTTNVAEIGPAVATLGRPLRLLRIPVDEWAVVLALALRAFP
MUL_1240|M.ulcerans_Agy99 LLALGAMVSWTTNVAEIGPAVATLGRPLRLLRIPVDEWAVVLALALRAFP
MLBr_00849|M.leprae_Br4923 LLGLGAMVSWTTNVAEIGPAMATLGRPLRIFRIPVDEWAVAVALALRAFP
MAV_3956|M.avium_104 LLGLGALVSWTTNVAQIAPAVAALGRPLRVLRIPVDDWSVALALALRTFP
MSMEG_4469|M.smegmatis_MC2_155 LLGLGALVSWTTNVADIAPAVATLGRPLGVFRIPVHDWAVTIALALRAFP
MAB_1714|M.abscessus_ATCC_1997 LLGTGAMVSWTTNVSEIAPAVAVLGRPLKVFGIPVDERAATLALALRSFP
Mflv_3462|M.gilvum_PYR-GCK AVALAAIVTTTTRTADMLDAATRAMTPLARFGFPVRQVAIALALTVRSIP
Mvan_3226|M.vanbaalenii_PYR-1 AVSLAAMVTLTTRTTDMIDAATRAMKPLARFGFPARQVGIALGLTIRSIP
TH_3631|M.thermoresistible__bu SVVLAAVVTVTTRVTAMLAALTALMRPLGRVGFPVDQLALALALTLRTIP
: .*:*: **..: : * : *: . :*. : . .:.*::*::*
Mb2352c|M.bovis_AF2122/97 MLIDEFQVLYAARRLRPKRMPPSRKARR-QRHARELIDLLAAAITVTLRR
Rv2325c|M.tuberculosis_H37Rv MLIDEFQVLYAARRLRPKRMPPSRKARR-QRHARELIDLLAAAITVTLRR
MMAR_3625|M.marinum_M MLIDEFQVLYAARRLRPRPMPVTRKARR-QRRGMEVIDLLAAAITVTLRR
MUL_1240|M.ulcerans_Agy99 MLIDEFQVLYAARRLRPRPMPVTRKARR-QRRGMEVIDLLAAAITVTLRR
MLBr_00849|M.leprae_Br4923 MLINEFQVLYAARRLRPKQILNSRKARR-QRQARELIDLLAAAITVTLRR
MAV_3956|M.avium_104 MLIDEFRVLYAARRLRPRRPAQTRWARL-RRPATDLIDVVVAVITVTLRR
MSMEG_4469|M.smegmatis_MC2_155 MLIDEFRTLYAARRLRPRERPDTFRARH-RQRVSGVVDLIAAAVTVALRR
MAB_1714|M.abscessus_ATCC_1997 MLSDEFRVLIAARRLRPPQVREGRRGRRGLRWIAEAVDMLTAAITAALRR
Mflv_3462|M.gilvum_PYR-GCK LLVEIIRQMEEARRARGLRISPR--------------IVFVPVIIGALRA
Mvan_3226|M.vanbaalenii_PYR-1 LLVEIIRQVDEARRARGLRVTPR--------------IVFVPVIVGALRA
TH_3631|M.thermoresistible__bu LMIDTVRQVEEARRARGLRFSPR--------------IVLAPVITAALDT
:: : .: : *** * :....: :*
Mb2352c|M.bovis_AF2122/97 ADEMGDAITARGGTGQLSAHPGRPKLADWVTLAITAMASGTAVAIESLIL
Rv2325c|M.tuberculosis_H37Rv ADEMGDAITARGGTGQLSAHPGRPKLADWVTLAITAMASGTAVAIESLIL
MMAR_3625|M.marinum_M ADEMGDAITARGGTGQLSANPTRPKLGDWVTLAITAATSGAAVVIETMLH
MUL_1240|M.ulcerans_Agy99 ADEMGDAITARGGTGQLSANPTRPKLGDWVTLAITAATSGAAVVIETMLH
MLBr_00849|M.leprae_Br4923 ADEMGDAITARGGIGQLSAYPARPKLADWVTLAITITTSSTAVVLETILH
MAV_3956|M.avium_104 ADEMGDAITARGGTGQISAAPSRPKRNDWIALSIASAVCAAAVAAELALL
MSMEG_4469|M.smegmatis_MC2_155 GDEMGDAITARGGTGQFSAAESKPRRLDWVAFAVLAVVCGTALALELTVL
MAB_1714|M.abscessus_ATCC_1997 ASEMGDAITARGGTGQISAHPARPGLNDAIALLLVIAVGTAVVLFEVL--
Mflv_3462|M.gilvum_PYR-GCK ADDFSEALIARGID------------------------------------
Mvan_3226|M.vanbaalenii_PYR-1 ADDLSEALIARGLD------------------------------------
TH_3631|M.thermoresistible__bu ADGFAEALTARGLD------------------------------------
.. :.:*: ***
Mb2352c|M.bovis_AF2122/97 HS-------
Rv2325c|M.tuberculosis_H37Rv HS-------
MMAR_3625|M.marinum_M AA-------
MUL_1240|M.ulcerans_Agy99 AA-------
MLBr_00849|M.leprae_Br4923 TAS------
MAV_3956|M.avium_104 AGH------
MSMEG_4469|M.smegmatis_MC2_155 GTSLPRRRG
MAB_1714|M.abscessus_ATCC_1997 ---------
Mflv_3462|M.gilvum_PYR-GCK ---------
Mvan_3226|M.vanbaalenii_PYR-1 ---------
TH_3631|M.thermoresistible__bu ---------