For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RAVKTVFDAPVDPALIDRSLAGAALGSVWLDIARPEFGTPGGPVTCDLLVVGGGYTGLWSALHAARRHPD RRIVLIEADRIGWAASGRNGGFVDASLTHGYENGKSRWPDELSTLEAMGIENLDGMQAEIAELGLDVEWE RTGMLAVATEPHQVEWLRDAAAQGQGTYLDQAQVRVEVNSPTYRAGLFSPDTCAIVNPARLVFELARACR DAGVEIYEHTRATRVDSGGAALRVHTDGPVVTCRQVILATNVFPSLLRRNRLHTVPVYDYVLATEPLTGE QLDRIGWRHRQGIGDSANQFHYYRLTADNRIVWGGYDAVYHFGRRVDPAYEDRAETYRRLAAHFFLTFPA LDDVRFTHRWAGAIDTNTRFCAHWGLAREGRVAYVNGFTGLGVGAARFAADVCLDLLDGTPTPRTRLNMV RRKPIPFPPEPLASVGIQATRWSLDRADHSAGRRNLLLRALDAAGLGFDS*
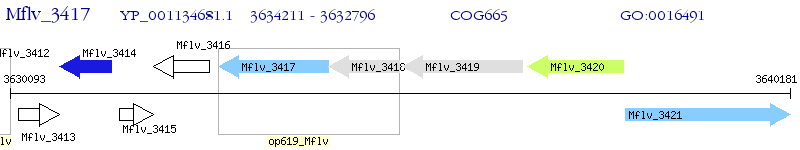
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3417 | - | - | 100% (471) | FAD dependent oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1109 | - | 7e-41 | 29.49% (434) | FAD dependent oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3531 | - | 3e-05 | 29.44% (197) | FAD dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2651c | - | 1e-28 | 27.66% (441) | oxidoreductase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2749 | - | 8e-05 | 26.24% (202) | FAD dependent oxidoreductase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3663 | - | 0.0 | 76.22% (471) | oxidoreductase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3170 | - | 0.0 | 87.29% (472) | FAD dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3417|M.gilvum_PYR-GCK -----MRAVKTVFDAPVDPALIDRSLAGAALGSVWLDIARPEFGTPGGP-
Mvan_3170|M.vanbaalenii_PYR-1 MPRRSVRAVQTVFDADVDPALVDRSLAGTTSGSVWLDIPRPQFPTPSGPP
MSMEG_3663|M.smegmatis_MC2_155 --------MQTVFDADVDPALLDRALAATTFGSMWLDVPRPEYPALTQP-
MAB_2651c|M.abscessus_ATCC_199 --------------------------MEYLFDTGWVDRPEVTAAALQGE-
MAV_2749|M.avium_104 --------------------------MTSLWMSERTERPWAASQLDQGP-
: : .
Mflv_3417|M.gilvum_PYR-GCK VTCDLLVVGGGYTGLWSALHAARRHPDRRIVLIEADRIGWAASGRNGGFV
Mvan_3170|M.vanbaalenii_PYR-1 VTCDLLVIGGGYTGLWSALHAARRHPDRRIVLIEADRIGWAASGRNGGFV
MSMEG_3663|M.smegmatis_MC2_155 VSCDLVVIGAGYTGLWTALHAAERHPDRRIVVIDSNRVGWAASGRNGGFV
MAB_2651c|M.abscessus_ATCC_199 IRCDVAVIGAGVGGMAAGLRLAERGVD--VVVLEAGICGWGASSRNGGYL
MAV_2749|M.avium_104 ASADVIVVGAGITGLMTAALLARAGKD--VLVLEARTVGACATANTTAKI
.*: *:*.* *: :. *. * :::::: * *:... . :
Mflv_3417|M.gilvum_PYR-GCK DASLTHGYENGKSRWPDELSTLEAMG-IENLDGMQAEIAELGLDVEWERT
Mvan_3170|M.vanbaalenii_PYR-1 DASLTHGYENGKSRWPNEIDTLEAMG-VENLDGMQAEIAELGLDVEWERT
MSMEG_3663|M.smegmatis_MC2_155 DASLTHGAENGKTRWPNEIGKLDALG-LENLDGMQADIERLGLDVEWQRT
MAB_2651c|M.abscessus_ATCC_199 TDALSGDPRMLSALHRRRLPGVMRFG-QSALRFAEDLIERHQIECDYNRV
MAV_2749|M.avium_104 SLLQGSQLSKVLPRHGREVTRAYVDGNREGQEWVLGHCEAHGVAVQRE--
. .: * . : : :
Mflv_3417|M.gilvum_PYR-GCK GMLAVATEPHQVEWLRDAAAQGQGTYLDQAQVR--VEVNSPTYRAGLFSP
Mvan_3170|M.vanbaalenii_PYR-1 GMLTVATEPHQVEWLADAAADGQGLFLDQPQVR--AEVDSPTYRAGLFSA
MSMEG_3663|M.smegmatis_MC2_155 GMLTVATEPHQRAWLAEAAAEGEGVFLDTAEVR--AQVNSPTYQAGLFSA
MAB_2651c|M.abscessus_ATCC_199 GWVSMASTTRHRAKNAERAAEVLRSLGADAEYVDGREFGLPHAFLGGVYE
MAV_2749|M.avium_104 ---DAYTYAQSVRGVPSARAEFEACQAVGLPVVWQDDAEVPFAYHGGVRL
: .: . *: : * * .
Mflv_3417|M.gilvum_PYR-GCK DTCAIVNPARLVFELARACRDAGVEIYEHTRATRVDSGGAALRVHTDGP-
Mvan_3170|M.vanbaalenii_PYR-1 DTCAIVHPAKLALELARACREAGVEVFEHTTATRIDSGGAALRVHTDGP-
MSMEG_3663|M.smegmatis_MC2_155 DTCAIVHPAKLALELARACTEAGVQIYEHTTARSINSGGAVLRIETGDTG
MAB_2651c|M.abscessus_ATCC_199 PHGGLCNAGKYALGLRKVLLASGARVYEQTPVQDLESSGSAVVITVPGG-
MAV_2749|M.avium_104 ADQAQFDPMPFLDSLAVELLGRGGRLVEHTRVRRVSWRGKGVRVHANQGS
. .. * * .: *:* . :. * : : .
Mflv_3417|M.gilvum_PYR-GCK -------VVTCRQVILAT-------NVFPSLLRRNR----LHTVP----V
Mvan_3170|M.vanbaalenii_PYR-1 -------AISCRQVVLAT-------NVFPSLLRRNR----LYTVP----V
MSMEG_3663|M.smegmatis_MC2_155 -----DTVITARQAVLAT-------NVFPSLLRRNR----FHTVP----V
MAB_2651c|M.abscessus_ATCC_199 -------KVRADRVLLAT-------NGYSADLPFAPP---RLAAP----L
MAV_2749|M.avium_104 DAAGHDVELHADQLVLATGIPILDRGGYFARVKPSRSYCLAFKVPGNITR
: . : :*** . : : : .*
Mflv_3417|M.gilvum_PYR-GCK YDYVLATEPLTGEQLDRIGWRHRQGIGDSANQFHYYRLTADNRIVWGGYD
Mvan_3170|M.vanbaalenii_PYR-1 YDYVLATEPLTETQLHRIGWQARQGIGDSANQFHYYRLTKDNRIVWGGYD
MSMEG_3663|M.smegmatis_MC2_155 YDYVLATEPLTPEQLDRIGWQGRQGVGDSANQFHYYRLTMDNRIVWGGYD
MAB_2651c|M.abscessus_ATCC_199 WVSLLETEPIDPERLDSTGWTNGAGFVSNNNIVSNFRKTPHNTIVFGTRN
MAV_2749|M.avium_104 PMMISTDSPTRSVRYAPVPDGERLIVGGAGHTVGREKSPSAALDELSAWT
: .* : . . : . : . .
Mflv_3417|M.gilvum_PYR-GCK AVYHFGRRVDPAYEDRAETYRRLAAHFFLTFPALDDVRFTHRWAGAIDTN
Mvan_3170|M.vanbaalenii_PYR-1 AVYHFGRRVNAAYEDRDETYRRLAAHFFITFPQLDDVRFTHRWAGAIDTN
MSMEG_3663|M.smegmatis_MC2_155 AIYHFGRKVKDAYEDRPATYRKLAAHFFLTFPQLDDVRFTHRWAGAIDTN
MAB_2651c|M.abscessus_ATCC_199 VQAPQG--PLGARTTDPAVAKDLLRGFREAFPSLRDVATQKTWGGWIGMS
MAV_2749|M.avium_104 RKHFPGAVQTHFWSAQDYTPIDHLPYVGPILPNSETIFVATGFNKWGMTN
* . . :* : : .
Mflv_3417|M.gilvum_PYR-GCK --TRFCAHWGLAREGRVAYVNGFTGLGVGAARFAADVCLDLLDGTPTPRT
Mvan_3170|M.vanbaalenii_PYR-1 --TRFCAHWGLAREGRVAYVNGYTGLGVAATRFAADVCLDLLDGAPSPRT
MSMEG_3663|M.smegmatis_MC2_155 --TRFCAHWGLARNGRVAYVNGFTGLGVAATRFAADVCLDLLDGRSTERT
MAB_2651c|M.abscessus_ATCC_199 --PTLLPVAGQS-SSRVLYTNGCNGYGMTQAPYLATLLADRLAG-DEMHE
MAV_2749|M.avium_104 GAAAALALSSRILGGRMDWARAFASWSPHELSGLATAVQANLEVGFNLTK
. . . .*: :... . . * *
Mflv_3417|M.gilvum_PYR-GCK RLNMVRRKPIPFPPEPLASVGIQATRWSLDR---ADHSAGRRNLLLRALD
Mvan_3170|M.vanbaalenii_PYR-1 ELEMVRSKPLPFPPEPLASIGIQATRWSLDR---ADHSAGRRNLLLRALD
MSMEG_3663|M.smegmatis_MC2_155 GLEMVRKRPLPFPPEPLASIGIQATRWSLDR---ADHSAGKRNFFLRALD
MAB_2651c|M.abscessus_ATCC_199 DLRAVWYDHNVFPPNPQFSALGARALWTLDR---ITDQVDR----LRS--
MAV_2749|M.avium_104 GWITPAVRIGRRSPVDGDGGVVSGPPWHLQARCRVDGTEHRVSPVCPHLG
.* . . * *: :
Mflv_3417|M.gilvum_PYR-GCK AAGLGFDS--------------------------------
Mvan_3170|M.vanbaalenii_PYR-1 AAGLGFDS--------------------------------
MSMEG_3663|M.smegmatis_MC2_155 TLGLGFDS--------------------------------
MAB_2651c|M.abscessus_ATCC_199 ----------------------------------------
MAV_2749|M.avium_104 GIVNWNDADKAWECPLHGSRFAPDGTLLEGPATRDLTASR