For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAVQEEDRVDALFHALSDRTRRDIMRRVLAGEHSVSTLASKYDMSFAAVQKHVAVLDRAGLITKRRRGRE QLASGDVAAVRSVASALEELEQIWRGRVERIDDLIATDKEPKNATKE*
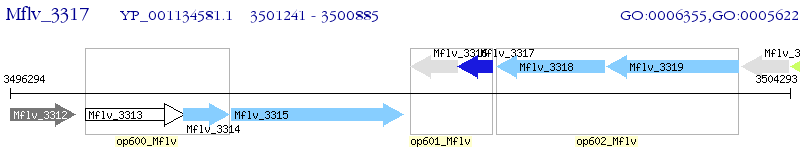
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3317 | - | - | 100% (118) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_3178 | - | 8e-09 | 39.29% (84) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_0605 | - | 8e-09 | 39.29% (84) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0591 | - | 1e-08 | 28.89% (90) | ArsR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0576 | - | 1e-08 | 28.89% (90) | ArsR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3862 | - | 8e-45 | 77.06% (109) | ArsR family transcriptional regulator |
| M. marinum M | MMAR_2816 | - | 8e-07 | 23.91% (92) | ArsR-type repressor |
| M. avium 104 | MAV_2783 | - | 2e-09 | 28.12% (96) | ArsR family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6762 | - | 4e-44 | 79.63% (108) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_3423 | - | 4e-10 | 29.46% (112) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_2937 | - | 8e-07 | 23.91% (92) | ArsR-type repressor |
| M. vanbaalenii PYR-1 | Mvan_3039 | - | 1e-48 | 81.36% (118) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3317|M.gilvum_PYR-GCK -MVAVQEEDRVDALFHALSDRTRRDIMRRVLAG-EHSVSTLASKYDMSFA
Mvan_3039|M.vanbaalenii_PYR-1 -MVIEHDEDRADALFHALADRTRRDIMRRVLAG-EHSVSALAAKYDMSFA
MAB_3862|M.abscessus_ATCC_1997 --MTDADEDKADAMFHALSDRTRRDILRRVLAG-EHSVSTLAANYDMSFA
MSMEG_6762|M.smegmatis_MC2_155 MWVTVIDEDRADALFHALADRTRRDIMRRVLAG-EQSVSALASKYDVSFA
Mb0591|M.bovis_AF2122/97 -------------MLEVAAEPTRRRLLQLLAPG-ERTVTQLASQFTVTRS
Rv0576|M.tuberculosis_H37Rv -------------MLEVAAEPTRRRLLQLLAPG-ERTVTQLASQFTVTRS
MMAR_2816|M.marinum_M ---MCTPERICDRIFVALASPVRRKLLEILTGQ-PLSAGELSDRFELSRP
MUL_2937|M.ulcerans_Agy99 ---MCTPERICDRIFVALASPVRRKLLEILTGR-PLSAGELSDRFELSRP
MAV_2783|M.avium_104 ---MVVVTGV-DRVFLALANPVRRELLQLLARH-PLSAGELSERFELSRP
TH_3423|M.thermoresistible__bu ----MTATDQLHPVFHALGDPNRMRLITRLCDAGPGSTSQVAQAVSVSRQ
:: . .. * :: : :. :: ::
Mflv_3317|M.gilvum_PYR-GCK AVQKHVAVLDRAGLITKRRRGREQLASGDVAAVRSV--------------
Mvan_3039|M.vanbaalenii_PYR-1 AVQKHVAVLDKAGLITKRRSGREQLASGDVEAVRSV--------------
MAB_3862|M.abscessus_ATCC_1997 AVQKHVAVLEKAGLLTKRRNGREQLASGDVEAVRSV--------------
MSMEG_6762|M.smegmatis_MC2_155 AVQKHVAVLEKAGLLTKRRRGREQLASGDVATVRSV--------------
Mb0591|M.bovis_AF2122/97 AISQHLGMLAEAGLVTARKQGRERYYRLDERGVLRLRALMESFWSDELDR
Rv0576|M.tuberculosis_H37Rv AISQHLGMLAEAGLVTARKQGRERYYRLDERGVLRLRALMESFWSDELDR
MMAR_2816|M.marinum_M AIAEHLKVLREAGLVSDEAQGRRRIYRLTAEPLAEL--------------
MUL_2937|M.ulcerans_Agy99 AIAEHLKVLREAGLVSDEAQGRRRIYRLTAEPLAEL--------------
MAV_2783|M.avium_104 AVAEHLKVLREAALVADERRGRQRIYHLTAEPLAEL--------------
TH_3423|M.thermoresistible__bu AATKHLLILEEAGLVRSDRRGRERIWRMQPEALDDA--------------
* :*: :* .*.*: **.: :
Mflv_3317|M.gilvum_PYR-GCK ------------ASALEELEQIWR---------------GRVERIDDLIA
Mvan_3039|M.vanbaalenii_PYR-1 ------------ASMLTELEQIWR---------------GRVSRIDELIA
MAB_3862|M.abscessus_ATCC_1997 ------------GAMLSELEQLWR---------------GRIARIDELIA
MSMEG_6762|M.smegmatis_MC2_155 ------------SQMLAELEQIWR---------------GRIARIDQLIS
Mb0591|M.bovis_AF2122/97 LVADAAHYPPSQGDCAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAA
Rv0576|M.tuberculosis_H37Rv LVADAAHYPPSQGDCAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAA
MMAR_2816|M.marinum_M ------------GEWLHPFEKLWR---------------DKLSALAETAE
MUL_2937|M.ulcerans_Agy99 ------------GEWLHPFEKLWR---------------EKLSALAEAAE
MAV_2783|M.avium_104 ------------GEWLHPFEKFWR---------------TRLRRLAEVAE
TH_3423|M.thermoresistible__bu ------------SEYLDRLARRWD---------------RALGRLRAFVE
. : : :
Mflv_3317|M.gilvum_PYR-GCK TDKEPKNATKE---------------------------------------
Mvan_3039|M.vanbaalenii_PYR-1 TDPRTENHDQEE--------------------------------------
MAB_3862|M.abscessus_ATCC_1997 RDRPSKD-------------------------------------------
MSMEG_6762|M.smegmatis_MC2_155 SEPLKED-------------------------------------------
Mb0591|M.bovis_AF2122/97 RIELRTGGAYRWTVTPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGS
Rv0576|M.tuberculosis_H37Rv RIELRTGGAYRWTVTPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGS
MMAR_2816|M.marinum_M EQQ-----------------------------------------------
MUL_2937|M.ulcerans_Agy99 EQQ-----------------------------------------------
MAV_2783|M.avium_104 ELE-----------------------------------------------
TH_3423|M.thermoresistible__bu APEQPDPGMSE---------------------------------------
Mflv_3317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3039|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3862|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6762|M.smegmatis_MC2_155 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 TVTITLTPVDGGTEVRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQHGDA
Rv0576|M.tuberculosis_H37Rv TVTITLTPVDGGTEVRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQRGDA
MMAR_2816|M.marinum_M --------------------------------------------------
MUL_2937|M.ulcerans_Agy99 --------------------------------------------------
MAV_2783|M.avium_104 --------------------------------------------------
TH_3423|M.thermoresistible__bu --------------------------------------------------
Mflv_3317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3039|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3862|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6762|M.smegmatis_MC2_155 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 GPDEWAAAPDPLDELSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQ
Rv0576|M.tuberculosis_H37Rv GPDEWAAAPDPLDELSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQ
MMAR_2816|M.marinum_M --------------------------------------------------
MUL_2937|M.ulcerans_Agy99 --------------------------------------------------
MAV_2783|M.avium_104 --------------------------------------------------
TH_3423|M.thermoresistible__bu --------------------------------------------------
Mflv_3317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3039|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3862|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6762|M.smegmatis_MC2_155 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 LADHLLRSLAIIGAAAGAQLAPRDVDAPLETQVADAAQAVMEAWRRRGLA
Rv0576|M.tuberculosis_H37Rv LADHLLRSLAIIGAAAGAQLAPRDVDAPLETQVADAAQAVMEAWRRRGLA
MMAR_2816|M.marinum_M --------------------------------------------------
MUL_2937|M.ulcerans_Agy99 --------------------------------------------------
MAV_2783|M.avium_104 --------------------------------------------------
TH_3423|M.thermoresistible__bu --------------------------------------------------
Mflv_3317|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3039|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3862|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6762|M.smegmatis_MC2_155 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 GTVELNSNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVSEYVLA
Rv0576|M.tuberculosis_H37Rv GTVELNSNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVSEYVLA
MMAR_2816|M.marinum_M --------------------------------------------------
MUL_2937|M.ulcerans_Agy99 --------------------------------------------------
MAV_2783|M.avium_104 --------------------------------------------------
TH_3423|M.thermoresistible__bu --------------------------------------------------
Mflv_3317|M.gilvum_PYR-GCK ------------------------------------------------
Mvan_3039|M.vanbaalenii_PYR-1 ------------------------------------------------
MAB_3862|M.abscessus_ATCC_1997 ------------------------------------------------
MSMEG_6762|M.smegmatis_MC2_155 ------------------------------------------------
Mb0591|M.bovis_AF2122/97 VAGKVITPATRNSAGFAAPAAVGSFAPVLDRLIAFTGRQPTAGHVSAT
Rv0576|M.tuberculosis_H37Rv VAGKVITPATRNSAGFAAPAAVGSFAPVLDRLIAFTGRQPTAGHVSAT
MMAR_2816|M.marinum_M ------------------------------------------------
MUL_2937|M.ulcerans_Agy99 ------------------------------------------------
MAV_2783|M.avium_104 ------------------------------------------------
TH_3423|M.thermoresistible__bu ------------------------------------------------