For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNKRDTCLAGNTSDLDAAVALFHSLSDATRLTIVRRLADGEARVVDLIGELGLAQSTVSAHVACLRDCGL VTGRPEGRQVFYSLSRPELLDVLAAAETLLAATGNAVALCPTYGTGARGAAIDTEETRQ
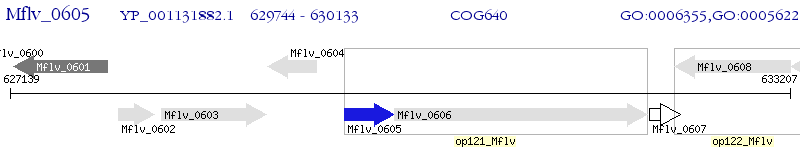
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0605 | - | - | 100% (129) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_3178 | - | 6e-69 | 100.00% (129) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_2865 | - | 4e-45 | 75.86% (116) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2017c | - | 5e-11 | 39.62% (106) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv1994c | - | 5e-11 | 39.62% (106) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4854c | - | 3e-37 | 76.00% (100) | ArsR family transcriptional regulator |
| M. marinum M | MMAR_2139 | - | 1e-41 | 75.70% (107) | transcriptional regulatory protein |
| M. avium 104 | MAV_5108 | - | 7e-09 | 32.91% (79) | transcriptional regulator, ArsR family protein |
| M. smegmatis MC2 155 | MSMEG_5603 | - | 2e-11 | 44.29% (70) | transcriptional regulator, ArsR family protein |
| M. thermoresistible (build 8) | TH_1240 | - | 2e-11 | 35.58% (104) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_3611 | - | 4e-08 | 34.15% (82) | ArsR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3640 | - | 2e-56 | 85.50% (131) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2017c|M.bovis_AF2122/97 ---------------------------------MLTCEMRESALARLGRA
Rv1994c|M.tuberculosis_H37Rv ---------------------------------MLTCEMRESALARLGRA
MSMEG_5603|M.smegmatis_MC2_155 ----------------------------------MQTALHTDALARFGHA
TH_1240|M.thermoresistible__bu --------------------------------VGMQTVTHTDALARFGHA
Mflv_0605|M.gilvum_PYR-GCK -----------------------MNKRDTCLAGNTS---DLDAAVALFHS
Mvan_3640|M.vanbaalenii_PYR-1 ---------------------MTMNKADGCLAGTTSGSSNLDAAVALFHS
MMAR_2139|M.marinum_M ---------------------MPMKIADDAPP-TASPREDLAGAVALFHS
MAB_4854c|M.abscessus_ATCC_199 -----------------------MTIKGSVVQSFAP--VSLQAASALFHS
MAV_5108|M.avium_104 --------------------------------MADSRQPLYRMKAEFFKT
MUL_3611|M.ulcerans_Agy99 MATSPSPSTAAHDDFVSRHEHDVGDFAEHPAFPAPPPREILEGAGELLRA
: ::
Mb2017c|M.bovis_AF2122/97 LADPTRCRILVALLDGVCYPGQLAAHLGLTRSNVSNHLSCLRGCGLVVAT
Rv1994c|M.tuberculosis_H37Rv LADPTRCRILVALLDGVCYPGQLAAHLGLTRSNVSNHLSCLRGCGLVVAT
MSMEG_5603|M.smegmatis_MC2_155 LSDVTRTRILLSLNESPNYPADLAEQLGVSRQTLSNHLACLRGCGLVVAV
TH_1240|M.thermoresistible__bu LSDATRARILLSLTKAPGYPAELADRLGVSRQVLSNHLACLRGCGLVVAV
Mflv_0605|M.gilvum_PYR-GCK LSDATRLTIVRRLADGEARVVDLIGELGLAQSTVSAHVACLRDCGLVTGR
Mvan_3640|M.vanbaalenii_PYR-1 LSDSARLAIVRRLADGEARVVDLIGELGLAQSTVSAHVACLRDCGLVTGR
MMAR_2139|M.marinum_M LSDPTRLAIARRLADGERRVVDLTRELGLPQSTVSSHLACLRDCGLIAGR
MAB_4854c|M.abscessus_ATCC_199 LSDETRLAILRRLACGEARVVDLTGELGLAQSTVSAHLGCLRECGLVDYR
MAV_5108|M.avium_104 LGHPARIRVLELLSSREYAVSEMLPEVGVEPANLSQQLSILRRAGLVTAR
MUL_3611|M.ulcerans_Agy99 LAAPVRIAIVLQLREAQRCVHELVDALGVPQPLVSQHLKILKAAGVVTGE
*. .* : * :: :*: :* :: *: .*::
Mb2017c|M.bovis_AF2122/97 YEGRQVRYALADSHLARALGELVQVVLAVDTDQPCVAERAASGEAVEMTG
Rv1994c|M.tuberculosis_H37Rv YEGRQVRYALADSHLARALGELVQVVLAVDTDQPCVAERAASGEAVEMTG
MSMEG_5603|M.smegmatis_MC2_155 PEGRRTRYELADARIGRALDDLMGLVLDVDPECRCVGPDGAVCGCG----
TH_1240|M.thermoresistible__bu PEGRRTRYELADARIGQALTDLMGLVLAVDPDC-CVGADGGRCSCV----
Mflv_0605|M.gilvum_PYR-GCK PEGRQVFYSLSRPELLDVLAAAETLLAATGNAVALCPTYGTGAR-GAAID
Mvan_3640|M.vanbaalenii_PYR-1 PEGRQVFYSLTRPELLDLLASAETLLAATGNAVALCPTYGTRARTGATTD
MMAR_2139|M.marinum_M PEGRQVFYALAVPDLLDLFAAAETVLAATGNAVTLCPNYGTRPTRASASR
MAB_4854c|M.abscessus_ATCC_199 PQGRSSVYRLARPEVLAVFTAAEDLLAATGNAVALCPNYG-HPDGHPETK
MAV_5108|M.avium_104 REGLSVRYALTSPQVAELLVVARTILTGVVAGQAETLEEIPESLLAEPTG
MUL_3611|M.ulcerans_Agy99 RSGREVLYRLVDQHLAHIVVDAVAHAGEDSA-------------------
.* * * : .
Mb2017c|M.bovis_AF2122/97 S-----------------
Rv1994c|M.tuberculosis_H37Rv S-----------------
MSMEG_5603|M.smegmatis_MC2_155 ------------------
TH_1240|M.thermoresistible__bu ------------------
Mflv_0605|M.gilvum_PYR-GCK TEETRQ------------
Mvan_3640|M.vanbaalenii_PYR-1 TKESL-------------
MMAR_2139|M.marinum_M RRGVKRAAPGVADWSPGR
MAB_4854c|M.abscessus_ATCC_199 RKVVR-------------
MAV_5108|M.avium_104 ------------------
MUL_3611|M.ulcerans_Agy99 ------------------