For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTPRERMVVSAALLIRERGAHPTAIADVLQHSGAPRGSAYHYFPGGRTQLLCEAVDYAAEFMAERLSRA SSSLRVLDDLFGYYRTQLQDSDFRAGCPVVAVAVESGDPDKPEQSAPVIERAAAALTRWSDIIEQRLIAD GVPRADAAALAMLVLTSFEGAIVVARATRDLKPLDLVAGQLASLIDARTVTRRK
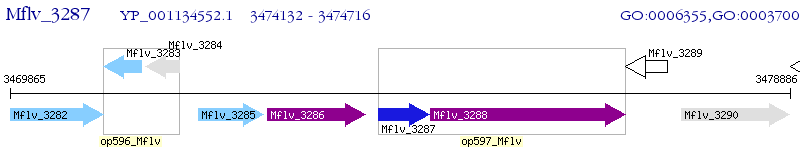
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3287 | - | - | 100% (194) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3237 | - | 8e-13 | 28.80% (184) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0202 | - | 4e-63 | 63.35% (191) | putative transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv0196 | - | 4e-63 | 63.35% (191) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1494 | - | 3e-61 | 62.37% (186) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0436 | - | 2e-61 | 60.75% (186) | transcriptional regulatory protein |
| M. avium 104 | MAV_4985 | - | 3e-64 | 67.03% (185) | regulatory protein, TetR |
| M. smegmatis MC2 155 | MSMEG_3520 | - | 6e-63 | 64.02% (189) | TetR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_3160 | - | 9e-30 | 40.00% (185) | PUTATIVE putative regulatory protein, TetR |
| M. ulcerans Agy99 | MUL_1086 | - | 2e-57 | 59.67% (181) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3009 | - | 1e-78 | 77.25% (189) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3287|M.gilvum_PYR-GCK -----------MTTPRERMVVSAALLIRERGAHPTAIADVLQHSGAPRGS
Mvan_3009|M.vanbaalenii_PYR-1 -----------MTSPRERMVISAALLIRERGAHPTAIADVLAHSGAPRGS
Mb0202|M.bovis_AF2122/97 -----------MQGPRERMVVSAALLIRERGAHATAISDVLQHSGAPRGS
Rv0196|M.tuberculosis_H37Rv -----------MQGPRERMVVSAALLIRERGAHATAISDVLQHSGAPRGS
MMAR_0436|M.marinum_M -----------MPSPRERMVISAVLLIRERGARATAISDVLQHSGAPRGS
MUL_1086|M.ulcerans_Agy99 ------------------MVISAVLLIRERGACATAISDVLQHSGAPRGS
MAV_4985|M.avium_104 -----------MRSPREKMVVSAALLIRERGAHATAISDVLEHSGAPRGS
MAB_1494|M.abscessus_ATCC_1997 -----------MSTPRERMVASAALLIRERGAHATAISDVLEHSGAPRGS
MSMEG_3520|M.smegmatis_MC2_155 -----------MASPRERMVRSAAVLIRERGAHPTAIADVLAHSGAPRGS
TH_3160|M.thermoresistible__bu MGVEVTRRRDPQSGPRAAMLRSAVALMRRQGVAATSFADVLADSGAPRGS
*: **. *:*.:*. .*:::*** .*******
Mflv_3287|M.gilvum_PYR-GCK AYHYFPGGRTQLLCEAVDYAA----EFMAERLSRASSSLRVLDDLFGYYR
Mvan_3009|M.vanbaalenii_PYR-1 AYHYFPGGRTQLLCEAVDYAG----EFIADRLAQASTTVEALDGLFDGYR
Mb0202|M.bovis_AF2122/97 AYHYFPGGRTQLLCEAVDYAG----EHVAAMINEAEGGLELLDALIDKYR
Rv0196|M.tuberculosis_H37Rv AYHYFPGGRTQLLCEAVDYAG----EHVAAMINEAEGGLELLDALIDKYR
MMAR_0436|M.marinum_M AYHYFPGGRTQLLCEAVDYAG----AHVAKIIGSANRSLDLLDTLIDQYR
MUL_1086|M.ulcerans_Agy99 AYHYFPGGRTQLLCEAVDYAG----AHVAKIIGSATRSLDLLDTLIDQYR
MAV_4985|M.avium_104 AYHYFPGGRTQLLCEAVDFAG----EYVAAVIAGAESGARLLDTLIDTYR
MAB_1494|M.abscessus_ATCC_1997 AYHYFPGGRTQLLSEAVDLAG----EYIAGMIDAKPSAVDALDILVASYR
MSMEG_3520|M.smegmatis_MC2_155 AYHYFPGGRTQLLCEAIDYAS----DHVTARLDRATDTFALLDDMVARFR
TH_3160|M.thermoresistible__bu IYHHFPAGKAQLIEEATRTAAAQLTERVARVLDEAADLPTALRELVGVWR
**:**.*::**: ** *. :: : * :. :*
Mflv_3287|M.gilvum_PYR-GCK TQLQDSDFRAGCPVVAVAVESGDPDKPEQSAPVIERAAAALTRWSDIIEQ
Mvan_3009|M.vanbaalenii_PYR-1 KQLRDSDFRAGCPVVAVTVEAGDPDKPEQSAAVIERAAAAFARWHDLLAQ
Mb0202|M.bovis_AF2122/97 QQLLSTDFRAGCPIAAVSVEAGDEQDRERMAPVIARAAAVFDRWSDLTAQ
Rv0196|M.tuberculosis_H37Rv QQLLSTDFRAGCPIAAVSVEAGDEQDRERMAPVIARAAAVFDRWSDLTAQ
MMAR_0436|M.marinum_M EQLRATDFRAGCPVAAVSVEAGEPSDRERMAPVVEHAAAVFDRWSDLIAQ
MUL_1086|M.ulcerans_Agy99 EQLRATDFRAGCPVAAVSVEAGEPSDRERMAPVVEHAAAVFDRWSDLIAQ
MAV_4985|M.avium_104 EQLRDSDFRAGCPVVAVAVEAGEQSDAER--PVIERAAAAFDRWTDLIAQ
MAB_1494|M.abscessus_ATCC_1997 KQLVRSDFRAGCPVVAVAVEAGDPESPASSQKLIEHSAAVFRRWIDLIAD
MSMEG_3520|M.smegmatis_MC2_155 DQLLASDFRAGCPVVAVAVEAGDPEKADSADAVRERAGAAFQRWIARITA
TH_3160|M.thermoresistible__bu DGLEAGDYAVGCPIVAAALGTER--------AARDVAGVAFADLTGLISD
* *: .***:.*.:: : :...:
Mflv_3287|M.gilvum_PYR-GCK RLIADGVPRADAAALAMLVLTSFEGAIVVARATRDLKPLDLVAGQLASLI
Mvan_3009|M.vanbaalenii_PYR-1 RLIADGLAQPAATALAMLVLTSFEGAIVVARASRDLAALDLVQEQLRELI
Mb0202|M.bovis_AF2122/97 RFIADGIPPDRAHELAVLATSTLEGAILLARVRRDLTPLDLVHRQLRNLL
Rv0196|M.tuberculosis_H37Rv RFIADGIPPDRAHELAVLATSTLEGAILLARVRRDLTPLDLVHRQLRNLL
MMAR_0436|M.marinum_M RFVSDGIPLDSAHELAVTAMSTLEGALLLARVRRDLTPLDVVHRQLRSML
MUL_1086|M.ulcerans_Agy99 RFVSDGIPLDSAHELAVTAMSTLEGALLLARVRRDLTPLYVVHRQLRSML
MAV_4985|M.avium_104 RFIADGIRRRRAAELAVLATSALEGAIVLARARRDPAPLDVVHRQLRDLL
MAB_1494|M.abscessus_ATCC_1997 KLVAVGVVRTRADELAILMTTSLEGAIIIARSTRDITPIDVVHRQLRTLI
MSMEG_3520|M.smegmatis_MC2_155 RLCDDGVPVERAEELATLITTSVEGAVLVARTTRDVKPLELVHRQLREQL
TH_3160|M.thermoresistible__bu KLVSDGMARRRANSVAVLILTAMEGALVLAQARRDPAALDAVVDELTLLC
:: *: * :* ::.***:::*: ** .: * :*
Mflv_3287|M.gilvum_PYR-GCK DART-----VTRRK----
Mvan_3009|M.vanbaalenii_PYR-1 RSHTS---PAARKRAPR-
Mb0202|M.bovis_AF2122/97 LAELP----ERSR-----
Rv0196|M.tuberculosis_H37Rv LAELP----ERSR-----
MMAR_0436|M.marinum_M LAQLP----EGNRNDHN-
MUL_1086|M.ulcerans_Agy99 LAQLP----EGNRNDHN-
MAV_4985|M.avium_104 AAELAGETAAGRKSRHDR
MAB_1494|M.abscessus_ATCC_1997 EEAGQ-------------
MSMEG_3520|M.smegmatis_MC2_155 TAATG----KKDRP----
TH_3160|M.thermoresistible__bu SGSAD-------------