For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFALATLLTLVNQVSGTPYVVGGDSPSGTDCSGLVSWVTNAATGRPVYGDRFHTGNIESALLARGFQYGT QPGALVVGWNSGHTAVTLPDGTPVSSGEGGGVRVGGGGAYQDQFTNHMFLPAPAAVPPPPDPFLSPPINQ LPPPPPPGAAPVVMMGHETALPPGAPLPPPPPGAPVPPPPPGMPVPPPPPGAPA
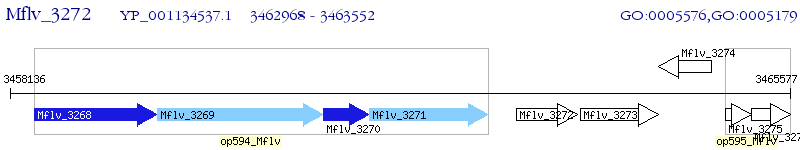
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3272 | - | - | 100% (194) | hypothetical protein Mflv_3272 |
| M. gilvum PYR-GCK | Mflv_1149 | - | 1e-12 | 39.67% (121) | hypothetical protein Mflv_1149 |
| M. gilvum PYR-GCK | Mflv_2951 | - | 5e-11 | 46.88% (64) | hypothetical protein Mflv_2951 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1145 | - | 1e-57 | 61.05% (190) | hypothetical protein Mb1145 |
| M. tuberculosis H37Rv | Rv1115 | - | 1e-57 | 61.05% (190) | hypothetical protein Rv1115 |
| M. leprae Br4923 | MLBr_02395 | Ag36 | 1e-10 | 37.78% (90) | proline rich antigenic protein |
| M. abscessus ATCC 19977 | MAB_4681c | - | 2e-51 | 61.35% (163) | hypothetical protein MAB_4681c |
| M. marinum M | MMAR_4345 | - | 5e-59 | 69.93% (153) | hypothetical protein MMAR_4345 |
| M. avium 104 | MAV_1247 | - | 2e-44 | 59.60% (151) | hypothetical protein MAV_1247 |
| M. smegmatis MC2 155 | MSMEG_3499 | - | 1e-72 | 65.52% (203) | hypothetical protein MSMEG_3499 |
| M. thermoresistible (build 8) | TH_1512 | - | 4e-33 | 77.92% (77) | POSSIBLE EXPORTED PROTEIN |
| M. ulcerans Agy99 | MUL_1750 | - | 1e-11 | 45.57% (79) | proline and glycine rich transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_2991 | - | 4e-89 | 79.70% (197) | hypothetical protein Mvan_2991 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3272|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2991|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3499|M.smegmatis_MC2_155 --------------------------------------------------
TH_1512|M.thermoresistible__bu --------------------------------------------------
MAB_4681c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1145|M.bovis_AF2122/97 -----------------------------------------MISTTRIDF
Rv1115|M.tuberculosis_H37Rv -----------------------------------------MISTTRIDF
MMAR_4345|M.marinum_M -----------------------------------------MISATNFDF
MAV_1247|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 MSQPPENPGNQADPQDGDQSTPGYPPPPNYGTPPPPPPPGYGAPPAGPPG
MLBr_02395|M.leprae_Br4923 --------MTDQPPPSGSNPTPAPPPPGSSG----GYEPSFAPSELGSAY
Mflv_3272|M.gilvum_PYR-GCK ----------MFALATLLTLVNQVSG------------------------
Mvan_2991|M.vanbaalenii_PYR-1 ------MSVRMFALATLLTLVNQVSG------------------------
MSMEG_3499|M.smegmatis_MC2_155 ----------MFALATLMALVQQVSG------------------------
TH_1512|M.thermoresistible__bu --------------------------------------------------
MAB_4681c|M.abscessus_ATCC_199 ----------MSDLAALLAFIHQVSG------------------------
Mb1145|M.bovis_AF2122/97 LWILSVAFAPMIALATLLTLINQVVG------------------------
Rv1115|M.tuberculosis_H37Rv LWILSVAFASMIALATLLTLINQVVG------------------------
MMAR_4345|M.marinum_M LWILSVASVLMIALATLLTLINQVSG------------------------
MAV_1247|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 YGAPPPPPGYGTPPPPPPPGYGQAPGGYAPPGYNPPPPPPPGYGPPPAGY
MLBr_02395|M.leprae_Br4923 PPPTAPPVGGSYPPPPPPGGSYPPP-------------------------
Mflv_3272|M.gilvum_PYR-GCK -TPYVVGGDSPSGTDCSGLVSWVTN--------------------AATGR
Mvan_2991|M.vanbaalenii_PYR-1 -TPYVSGGDSPAGTDCSGLVSWVTN--------------------AATGR
MSMEG_3499|M.smegmatis_MC2_155 -TPYVSGGDSPSGTDCSGLASWVSN--------------------VATGR
TH_1512|M.thermoresistible__bu ----------------------VAN--------------------VATGR
MAB_4681c|M.abscessus_ATCC_199 -TPYISGGDTARGTDCSGLASWVAN--------------------VATDR
Mb1145|M.bovis_AF2122/97 -TPYIPGGDSPAGTDCSELASWVSN--------------------AATAR
Rv1115|M.tuberculosis_H37Rv -TPYIPGGDSPAGTDCSELASWVSN--------------------AATAR
MMAR_4345|M.marinum_M -TPYIVGGDSPAGTDCSGLASWVSN--------------------AATDR
MAV_1247|M.avium_104 --------------------------------------------------
MUL_1750|M.ulcerans_Agy99 STQPGFGGPAKPPFSVGEAISWAWNRFTQNAMALVVPIVIYGLILSAVGG
MLBr_02395|M.leprae_Br4923 --PPPGGSYPPPPPSTGAYAPPPPG--------------------PAIRS
Mflv_3272|M.gilvum_PYR-GCK PVYGDRFHTGNIESALLARGFQYGTQPG--------------------AL
Mvan_2991|M.vanbaalenii_PYR-1 PVYGDRFHTGNIESALLARGFQYGTQPG--------------------AL
MSMEG_3499|M.smegmatis_MC2_155 PAFGDRFHTGNQEQALLARGFKYGTQPG--------------------AL
TH_1512|M.thermoresistible__bu PAFGDRFHTGNQEAALLARGFKYGTAPG--------------------AL
MAB_4681c|M.abscessus_ATCC_199 PAFGSRFNTGNQERALLARGFKYGTAPG--------------------AL
Mb1145|M.bovis_AF2122/97 PVFGDRFNTGNEEAALAARGFQQGTAPN--------------------AL
Rv1115|M.tuberculosis_H37Rv PVFGDRFNTGNEEAALAARGFQQGTAPN--------------------AL
MMAR_4345|M.marinum_M PIFGDRFNTGNEEAALAARGFQHGTAPN--------------------AL
MAV_1247|M.avium_104 -MFGDRFNTGNEEAALLARGFQYGTAPN--------------------AL
MUL_1750|M.ulcerans_Agy99 VMVGLFFAFSDRTSTTYTDAYGNTSETVNMTMSPLASIVMFIGYLAVFAV
MLBr_02395|M.leprae_Br4923 LPKEAYTFWVTRVLAYVIDNIPATVLLG-------------------IGM
: .:
Mflv_3272|M.gilvum_PYR-GCK VVGWNSGHTA--VTLPDGTPVSSG----------------------EGGG
Mvan_2991|M.vanbaalenii_PYR-1 VVGWNRGHTA--VTLPDGTPVSSG----------------------EGGG
MSMEG_3499|M.smegmatis_MC2_155 VIGWNSGHTA--VTLPDGTPVSSG----------------------EGGG
TH_1512|M.thermoresistible__bu VIGWNSTHTA--VTLPDGTPVSAG----------------------EGGG
MAB_4681c|M.abscessus_ATCC_199 VIGWNGGHTA--VTLPDGTAVSSG----------------------EGGG
Mb1145|M.bovis_AF2122/97 VIGWNGHHTA--VTLPDGTPVSSG----------------------EGGG
Rv1115|M.tuberculosis_H37Rv VIGWNGHHTA--VTLPDGTPVSSG----------------------EGGG
MMAR_4345|M.marinum_M VIGWNGRHTA--VTLPDGTSVSSG----------------------EGGG
MAV_1247|M.avium_104 VIGWNGGHTA--VTLPDGTAVSSG----------------------EHGG
MUL_1750|M.ulcerans_Agy99 VLFMHAGITTGCLDIADGKPVTIGSFFKPRNLGMVILTGLLVIVLTAIGS
MLBr_02395|M.leprae_Br4923 LIQTLTKQEACVTDITQYNVNQYC----------------------ATQP
:: : :.: .
Mflv_3272|M.gilvum_PYR-GCK VRVGGGGAYQDQFTNHMFLPAPAAVPPPPDPFLSPPINQLPPPP------
Mvan_2991|M.vanbaalenii_PYR-1 VKIGGGGAYQEQFTNHMFLPAPAEAVPAPDPVLAPPMSPFPPPPG-----
MSMEG_3499|M.smegmatis_MC2_155 VKIGGGGAYQPQFNHHMYLPAEEVQPDAGLPV-GAPPPPEASLPPLPPPP
TH_1512|M.thermoresistible__bu VKIGGGGAFXXPFG------------------------------------
MAB_4681c|M.abscessus_ATCC_199 VRIGGGGAYQPQFKRHMYLPMDSDDAAGSAPL-DVPADAEG---------
Mb1145|M.bovis_AF2122/97 VRVGGGGAYQPKFTHHMYLPMDVDAGEDQPPAPDEPVTAVDDVEP-----
Rv1115|M.tuberculosis_H37Rv VRVGGGGAYQPKFTHHMYLPMDVDAGEDQPPAPDEPVTAVDDVEP-----
MMAR_4345|M.marinum_M VRVGGGGAYQAQFTHHMYLPMDEDMVDVDLPAPEDPPAVEDVVNP-----
MAV_1247|M.avium_104 VHVGGAGAYQAGFTHHMFLPMPPEDGGAPPPAMFGPPPPPDAPPP-----
MUL_1750|M.ulcerans_Agy99 VLCIVPGLIFGFLAQFAIIAAVDRSLSPIDSIKSSCATVRAELGNTALSW
MLBr_02395|M.leprae_Br4923 TGIGMLAFWFAWLMATAYLVWNYGYRQGATGSSIGKTVMKFKVIS-----
. . :
Mflv_3272|M.gilvum_PYR-GCK -------------------PPGAAPVVMMGHETALPPGAPLPPPPPGAPV
Mvan_2991|M.vanbaalenii_PYR-1 ------------AVPPPPPPPPAGPVVLMGQQAPVPP-------PPGAPV
MSMEG_3499|M.smegmatis_MC2_155 ADPMAPAPPMAGPPPVDPFAPPPPPEPAVVPVVLDVP-------APPAPE
TH_1512|M.thermoresistible__bu --------------------------------------------------
MAB_4681c|M.abscessus_ATCC_199 --------------------PEVIPASVLVPQPESVP-------ADEAPA
Mb1145|M.bovis_AF2122/97 ----------EMPAPCPTQRPPVTPRHNLCNKLRTMPGALSAALAAAAPV
Rv1115|M.tuberculosis_H37Rv ----------EMPAPCPTQRPPVTPRHNLCNKLRTMPGALSAALAAAAPV
MMAR_4345|M.marinum_M ----------DELPP-----PPEDP----------------------GPV
MAV_1247|M.avium_104 -------------PPDAPAPPPDAPPAPPEAPAVLID--------APAPV
MUL_1750|M.ulcerans_Agy99 LVQYAVVLAGELLCFVGMLVGVPVASLIQTYTWRKLSGGQVVELSQPGPP
MLBr_02395|M.leprae_Br4923 -------------EATGQPIGFGMSVVRQLAHFVDAVICCIGFLFPLWDS
Mflv_3272|M.gilvum_PYR-GCK PPPPPGMPVPPPPPGAPA---------
Mvan_2991|M.vanbaalenii_PYR-1 PPPP-GAPVPPAP--------------
MSMEG_3499|M.smegmatis_MC2_155 APPALGV--------------------
TH_1512|M.thermoresistible__bu ---------------------------
MAB_4681c|M.abscessus_ATCC_199 QVAPVEI--------------------
Mb1145|M.bovis_AF2122/97 WPAPISGCRGFSTSLLAKRNHPVIVGK
Rv1115|M.tuberculosis_H37Rv WPAPISGCRGFSTSLLAKRNHPVIVGK
MMAR_4345|M.marinum_M DDLEISNT-------------------
MAV_1247|M.avium_104 PPPPPGP--------------------
MUL_1750|M.ulcerans_Agy99 AGLPPGPPPGAPPGPQFG---------
MLBr_02395|M.leprae_Br4923 KRQTLADKIMTTVCLPI----------