For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRYHRRMAMTTDDCGSVRPSTNLSPAAALFHGLADPTRLAILQRLADGEARVVDLTGLLSMAQSTVSAHL ACLRDCGLVVGRPEGRQMFYSLTRPELLDLLASAETLLAATGNAVALCPNYGTRSRTGATIDAEEIGR
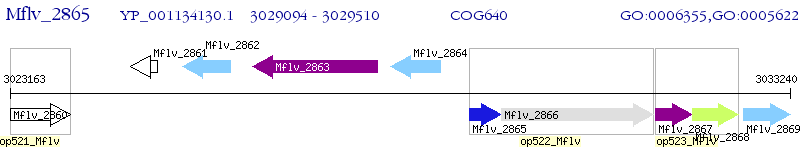
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2865 | - | - | 100% (138) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_3178 | - | 4e-45 | 75.86% (116) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_0605 | - | 4e-45 | 75.86% (116) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2017c | - | 1e-11 | 43.68% (87) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv1994c | - | 1e-11 | 43.68% (87) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4854c | - | 2e-35 | 71.29% (101) | ArsR family transcriptional regulator |
| M. marinum M | MMAR_2139 | - | 2e-41 | 67.80% (118) | transcriptional regulatory protein |
| M. avium 104 | MAV_5108 | - | 2e-08 | 34.15% (82) | transcriptional regulator, ArsR family protein |
| M. smegmatis MC2 155 | MSMEG_5603 | - | 7e-10 | 48.39% (62) | transcriptional regulator, ArsR family protein |
| M. thermoresistible (build 8) | TH_1240 | - | 8e-10 | 36.67% (90) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_3287 | - | 9e-09 | 39.24% (79) | ArsR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3640 | - | 6e-50 | 76.98% (126) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2017c|M.bovis_AF2122/97 -------------------MLTCEMRESALARLGRALADPTRCRILVALL
Rv1994c|M.tuberculosis_H37Rv -------------------MLTCEMRESALARLGRALADPTRCRILVALL
MSMEG_5603|M.smegmatis_MC2_155 --------------------MQTALHTDALARFGHALSDVTRTRILLSLN
TH_1240|M.thermoresistible__bu ------------------VGMQTVTHTDALARFGHALSDATRARILLSLT
Mflv_2865|M.gilvum_PYR-GCK -MRYHRRMAMTTDD---CGSVRPSTNLSPAAALFHGLADPTRLAILQRLA
Mvan_3640|M.vanbaalenii_PYR-1 -------MTMNKADGCLAGTTSGSSNLDAAVALFHSLSDSARLAIVRRLA
MMAR_2139|M.marinum_M -------MPMKIADD-APPTASPREDLAGAVALFHSLSDPTRLAIARRLA
MAB_4854c|M.abscessus_ATCC_199 ---------MTIKGS-VVQSFAPVS-LQAASALFHSLSDETRLAILRRLA
MAV_5108|M.avium_104 ------------------MADSRQPLYRMKAEFFKTLGHPARIRVLELLS
MUL_3287|M.ulcerans_Agy99 MSNRVEMDAPCCVTAPLLREPLEASSAVEMAARFKALADPVRLQLLSSVA
: *.. .* : :
Mb2017c|M.bovis_AF2122/97 ---DGVCYPGQLAAHLGLTRSNVSNHLSCLRGCGLVVATYEGRQVRYALA
Rv1994c|M.tuberculosis_H37Rv ---DGVCYPGQLAAHLGLTRSNVSNHLSCLRGCGLVVATYEGRQVRYALA
MSMEG_5603|M.smegmatis_MC2_155 ---ESPNYPADLAEQLGVSRQTLSNHLACLRGCGLVVAVPEGRRTRYELA
TH_1240|M.thermoresistible__bu ---KAPGYPAELADRLGVSRQVLSNHLACLRGCGLVVAVPEGRRTRYELA
Mflv_2865|M.gilvum_PYR-GCK ---DGEARVVDLTGLLSMAQSTVSAHLACLRDCGLVVGRPEGRQMFYSLT
Mvan_3640|M.vanbaalenii_PYR-1 ---DGEARVVDLIGELGLAQSTVSAHVACLRDCGLVTGRPEGRQVFYSLT
MMAR_2139|M.marinum_M ---DGERRVVDLTRELGLPQSTVSSHLACLRDCGLIAGRPEGRQVFYALA
MAB_4854c|M.abscessus_ATCC_199 ---CGEARVVDLTGELGLAQSTVSAHLGCLRECGLVDYRPQGRSSVYRLA
MAV_5108|M.avium_104 ---SREYAVSEMLPEVGVEPANLSQQLSILRRAGLVTARREGLSVRYALT
MUL_3287|M.ulcerans_Agy99 SHAGGEACVCDISAGVEVGQPTISHHLKVLRDAGLLTSQRRASWVYYAVV
:: : : :* :: ** .**: .. * :.
Mb2017c|M.bovis_AF2122/97 DSHLARALGELVQVVLAVDTDQPCVAERAASGEAVEMTGS----------
Rv1994c|M.tuberculosis_H37Rv DSHLARALGELVQVVLAVDTDQPCVAERAASGEAVEMTGS----------
MSMEG_5603|M.smegmatis_MC2_155 DARIGRALDDLMGLVLDVDPECRCVGPDGAVCGCG---------------
TH_1240|M.thermoresistible__bu DARIGQALTDLMGLVLAVDPDC-CVGADGGRCSCV---------------
Mflv_2865|M.gilvum_PYR-GCK RPELLDLLASAETLLAATGNAVALCPNYGTRSRTGATIDAEEIGR-----
Mvan_3640|M.vanbaalenii_PYR-1 RPELLDLLASAETLLAATGNAVALCPTYGTRARTGATTDTKESL------
MMAR_2139|M.marinum_M VPDLLDLFAAAETVLAATGNAVTLCPNYGTRPTRASASRRRGVKRAAPGV
MAB_4854c|M.abscessus_ATCC_199 RPEVLAVFTAAEDLLAATGNAVALCPNYG-HPDGHPETKRKVVR------
MAV_5108|M.avium_104 SPQVAELLVVARTILTGVVAGQAETLEEIPESLLAEPTG-----------
MUL_3287|M.ulcerans_Agy99 -PEALNALSRLLSVHAGSATELETSA------------------------
. : :
Mb2017c|M.bovis_AF2122/97 -------
Rv1994c|M.tuberculosis_H37Rv -------
MSMEG_5603|M.smegmatis_MC2_155 -------
TH_1240|M.thermoresistible__bu -------
Mflv_2865|M.gilvum_PYR-GCK -------
Mvan_3640|M.vanbaalenii_PYR-1 -------
MMAR_2139|M.marinum_M ADWSPGR
MAB_4854c|M.abscessus_ATCC_199 -------
MAV_5108|M.avium_104 -------
MUL_3287|M.ulcerans_Agy99 -------