For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TIHGATGNALDVTGLTKAFGSHTVLHGIDLGVTAGTITAVVGASGCGKTTLLRLVAGFENPDAGTVSIAG RQVAAAGSSIPAHRRDVGYVAQDGALFPHLTVGQNIAYGLSGRRNVQPRVAELLATVSLDESYAARRPHE LSGGQQQRVALARALARQPALMLLDEPFSALDTGLRASTRKAVATLLSDAGVTTLIVTHDQEEALSIADQ VAVMRDGRFTKVGTPQAVYREPNDRFTAEFLGDCVLLPCTVTGGVATSALGRLPVRGTVPDGGGTLMLRP EQLEAAAVSDDDRSDGMATVLASEFRGHDVLLTVDLGGDAAPITVRQHSVAPPAVDAKVRIHVTGAAVVF GDRPRDGR*
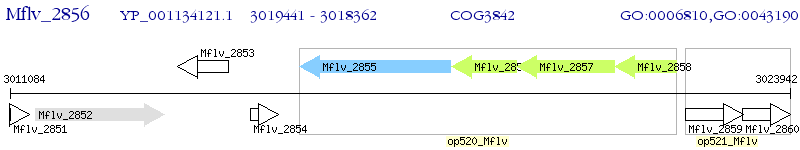
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2856 | - | - | 100% (359) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_3361 | - | 1e-79 | 48.39% (341) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_3778 | - | 3e-58 | 42.04% (314) | ABC transporter related |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 4e-52 | 45.38% (238) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 4e-52 | 45.38% (238) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01424 | - | 2e-46 | 35.31% (337) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_1655 | - | 8e-55 | 46.03% (239) | sulfate ABC transporter, ATP-binding protein SysA |
| M. marinum M | MMAR_3715 | cysA1 | 7e-53 | 44.94% (247) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 3e-52 | 45.38% (238) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. smegmatis MC2 155 | MSMEG_3633 | - | 4e-80 | 51.04% (335) | ferric cations import ATP-binding protein FbpC |
| M. thermoresistible (build 8) | TH_3667 | - | 9e-81 | 48.31% (354) | PUTATIVE ferric cations import ATP-binding protein FbpC |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 5e-53 | 44.94% (247) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_3091 | - | 3e-81 | 48.84% (346) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3633|M.smegmatis_MC2_155 --------------------MP-----DPAGISVR-------GVHKSYRD
Mvan_3091|M.vanbaalenii_PYR-1 --------------------MPGCAVTDAAAVSAR-------GIHKAFGG
TH_3667|M.thermoresistible__bu VIKTMKTKPADTSGAGPAVSNPAVSNPDVSAPQLRGADVEVQGLHKSFGA
Mflv_2856|M.gilvum_PYR-GCK ---------------------MTIHGATGNALDVT-------GLTKAFGS
Mb2419c|M.bovis_AF2122/97 -------------------MT--------YAIVVA-------DATKRYGD
Rv2397c|M.tuberculosis_H37Rv -------------------MT--------YAIVVA-------DATKRYGD
MAV_1785|M.avium_104 -------------------MTDAGTDRGDIAITVR-------DAYKRYGD
MMAR_3715|M.marinum_M -------------MKKETSLTGPG--TGGHAIVVR-------DAFKQYGD
MUL_3658|M.ulcerans_Agy99 -------------------MTGPG--TGGHAIVVR-------DAFKQYGD
MAB_1655|M.abscessus_ATCC_1997 -------------------MTAEN------TITVR-------GANKHYGD
MLBr_01424|M.leprae_Br4923 ------------------------MASVTFEQATR---------QFPDTD
MSMEG_3633|M.smegmatis_MC2_155 RRVLAGVDLDVAAGTITAVLGPSGCGKTTLLRIMAGFDDPDAGVVRIAGH
Mvan_3091|M.vanbaalenii_PYR-1 RTVLCGVDLEMAAGSITAVLGPSGCGKTTLLRILAGFEDPDSGTVRIAGE
TH_3667|M.thermoresistible__bu KAVLRGVDLTVSAGTLTAVLGPSGCGKTTLLRILAGFDDPDAGTVRIGGH
Mflv_2856|M.gilvum_PYR-GCK HTVLHGIDLGVTAGTITAVVGASGCGKTTLLRLVAGFENPDAGTVSIAGR
Mb2419c|M.bovis_AF2122/97 FVALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTITINGR
Rv2397c|M.tuberculosis_H37Rv FVALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTITINGR
MAV_1785|M.avium_104 FVALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDTGTVTIYGR
MMAR_3715|M.marinum_M FVALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDSGNITINGR
MUL_3658|M.ulcerans_Agy99 FVALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDSGNITINGR
MAB_1655|M.abscessus_ATCC_1997 FAALDNIDFDVPSGSLTALLGPSGSGKSTLLRSIAGLDNPDSGTITINGR
MLBr_01424|M.leprae_Br4923 RPALDRLDLDVDDGEFVVVVGPSGCGKTTSLRMVAGLETLDSGCIRIGDR
.* :*: : * :..::*.**.**:* ** :**:: *:* : * ..
MSMEG_3633|M.smegmatis_MC2_155 TVVGDGPVVPAHRRRVGLMPQEGALFPHLSVGGNVAFGLP----SRNGVA
Mvan_3091|M.vanbaalenii_PYR-1 TVAGDGAAVPVHRRRVGLMPQEGALFPHLSVGENVAFGLGR--ADRATAS
TH_3667|M.thermoresistible__bu TLAGGGAPVPAHRRRVGLMPQEGALFPHLSVAENIAFGLRR--TRRAGRA
Mflv_2856|M.gilvum_PYR-GCK QVAAAGSSIPAHRRDVGYVAQDGALFPHLTVGQNIAYGLS----GRRNVQ
Mb2419c|M.bovis_AF2122/97 DVTR----VPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRKRPKAEIK
Rv2397c|M.tuberculosis_H37Rv DVTR----VPPQRRGIGFVFQHYAAFKHLTVRDNVAFGLKIRKRPKAEIK
MAV_1785|M.avium_104 DVTR----VPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKVRKRPKAEIK
MMAR_3715|M.marinum_M DVTR----VPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAEIR
MUL_3658|M.ulcerans_Agy99 DVTR----VPPQRRGIGFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAEIR
MAB_1655|M.abscessus_ATCC_1997 DVTG----VPPQRRDIGFVFQHYAAFKHLTVWENVAFGLKIRKKPKAEIK
MLBr_01424|M.leprae_Br4923 DVTH----VDPKDRDVAMVFQNYALYPHMTVAQNLGFALKISKISKAEIH
:. : : * :. : *. * : *::* *:.:.* :
MSMEG_3633|M.smegmatis_MC2_155 ADVAHWLEVVGLPG-LADARPHELSGGQQQRVALARALAAKPRVLLLDEP
Mvan_3091|M.vanbaalenii_PYR-1 AQVAHWLDVVGLEG-LADARPHEISGGQQQRVALARALAARPRVLLLDEP
TH_3667|M.thermoresistible__bu DTVRYWLEVVGLEG-AAEARPHELSGGQQQRVALARALAAEPRVLLLDEP
Mflv_2856|M.gilvum_PYR-GCK PRVAELLATVSLDESYAARRPHELSGGQQQRVALARALARQPALMLLDEP
Mb2419c|M.bovis_AF2122/97 AKVDNLLQVVGLSG-FQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEP
Rv2397c|M.tuberculosis_H37Rv AKVDNLLQVVGLSG-FQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEP
MAV_1785|M.avium_104 AKVDNLLEVVGLSG-FQGRYPNQLSGGQRQRMALARALAVDPQVLLLDEP
MMAR_3715|M.marinum_M DKVDNLLEVVGLSG-FHGRYPNQLSGGQRQRMALARALAVDPEVLLLDEP
MUL_3658|M.ulcerans_Agy99 DKVDNLLEVVGLSG-FHGRYPDQLSGGQRQRMALARALAVDPEVLLLDEP
MAB_1655|M.abscessus_ATCC_1997 KKVDDLLEVVGLSG-FHTRYPAQLSGGQRQRMALARALAVDPQVLLLDEP
MLBr_01424|M.leprae_Br4923 DRILVVAKLLDLQP-YLDRKPKDLSGGQRQRVAMGRAIVRRPQVFLMDEP
: :.* * ::****:**:*:.**:. * ::*:***
MSMEG_3633|M.smegmatis_MC2_155 FAALDAGLRVRVREDIAEILRAAGTTAVLVTHDQAEALSLADSVALLFDG
Mvan_3091|M.vanbaalenii_PYR-1 FAALDAGLRVRVREDIATILRDTGTTALLVTHDQSEALSLADSVALLIDG
TH_3667|M.thermoresistible__bu FVALDAGLRVRVREEIAAILRAAGTTAVLVTHDQSEALSLADSVALLMDG
Mflv_2856|M.gilvum_PYR-GCK FSALDTGLRASTRKAVATLLSDAGVTTLIVTHDQEEALSIADQVAVMRDG
Mb2419c|M.bovis_AF2122/97 FGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHKG
Rv2397c|M.tuberculosis_H37Rv FGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHKG
MAV_1785|M.avium_104 FGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNQG
MMAR_3715|M.marinum_M FGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNSG
MUL_3658|M.ulcerans_Agy99 FGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNSG
MAB_1655|M.abscessus_ATCC_1997 FGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNKG
MLBr_01424|M.leprae_Br4923 LSNLDAKLRAQTRNQIAELQCQLGTTTVYVTHDQVEAMTMGDRVAVLCYG
: **: :* * : : .*:: ***** **: :.* :*:: *
MSMEG_3633|M.smegmatis_MC2_155 TVAQHAAPTDLYDRPANLQTARFVG--ATVELAGTCGDGVARTALGAHPV
Mvan_3091|M.vanbaalenii_PYR-1 VIAQHGTPADLYERPGTLANARFIG--STVEIAGTADAGTVHTSLGPLRT
TH_3667|M.thermoresistible__bu RLAQVGAPADIYERPATPAAARFVG--ATVELPGQARDGVVHTALGSHPA
Mflv_2856|M.gilvum_PYR-GCK RFTKVGTPQAVYREPNDRFTAEFLG--DCVLLPCTVTGGVATSALGRLPV
Mb2419c|M.bovis_AF2122/97 RIEQVGSPTDVYDAPANAFVMSFLG--AVSTLNGSLVRPHDIRVGRTPNM
Rv2397c|M.tuberculosis_H37Rv RIEQVGSPTDVYDAPANAFVMSFLG--AVSTLNGSLVRPHDIRVGRTPNM
MAV_1785|M.avium_104 RIEQIGSPTEVYDAPTNAFVMSFLG--AVSTLNGTLVRPHDIRVGRTPEM
MMAR_3715|M.marinum_M RIEQVGSPTEVYDAPANAFVMSFLG--AVSALNGALVRPHDIRVGRTPEM
MUL_3658|M.ulcerans_Agy99 RIEQVGSPTEVYDAPTNAFVMSFLG--AVSALNGALVRPHDIRVGRTPEM
MAB_1655|M.abscessus_ATCC_1997 RIEQVGSPEDLYDRPANSFVMSFLG--PVAKLNGILVRPHDIRVGRNADL
MLBr_01424|M.leprae_Br4923 VLQQFATPRELYHKPENVFVAGFIGSPGMNLVTLSIVDSSVLLGDWPIRI
. : .:* :* * *:* :
MSMEG_3633|M.smegmatis_MC2_155 RVRVPDGPVVVVLRPEQLR--AAEAGTPGGDHDAVVRACRFYGADTVLHV
Mvan_3091|M.vanbaalenii_PYR-1 RGPVADGPAVVALRPEQLRFGTGLSGTPG-----RVTARRFYGADTVVHV
TH_3667|M.thermoresistible__bu RLPVPDGPVVVVLRPEELRLERSRDGVPA----AAVQSVRFYGPQSAVQV
Mflv_2856|M.gilvum_PYR-GCK RGTVPDGGGTLMLRPEQLEAAAVSDDDRS-DGMATVLASEFRGHDVLLTV
Mb2419c|M.bovis_AF2122/97 AVAAADGTAGSTGVLRAVVDRVVVLGFEVR----VELTSAATGGAFTAQI
Rv2397c|M.tuberculosis_H37Rv AVAAADGTAGSTGVLRAVVDRVVVLGFEVR----VELTSAATGGAFTAQI
MAV_1785|M.avium_104 AVAAEDGTAESTGVARAIVDRVVKLGFEVR----VELTSAATGGPFTAQI
MMAR_3715|M.marinum_M AVAAADGTGESTGVTRAVVDRIVVLGFEVR----VELTSAATGGAFTAQI
MUL_3658|M.ulcerans_Agy99 AVAAADGTGESTGVTRAVVDRIVVLGFEVR----VELTSAATGGAFTAQI
MAB_1655|M.abscessus_ATCC_1997 ARAAAEGTGETTGVTKATVDRVVYLGFEVR----VELTSAATGEQFTAQI
MLBr_01424|M.leprae_Br4923 PREIASAASEVIIGVRPEHFELGSLGVEVEIDMVEELGADAYLYGRIAGA
.. . .
MSMEG_3633|M.smegmatis_MC2_155 VLR-DGTAVRLRCAGDARVEAGTVVSVTVDG---PVLAYPSPAGG-----
Mvan_3091|M.vanbaalenii_PYR-1 ALT-DGTALRLRSPVPTGLDDGSPVTIEVAG---DVLAYPSPTSGQRVPL
TH_3667|M.thermoresistible__bu RLA-DDTVVTVLTAGDAGVSVGEPVGVRVDG---SVLAYPAPGRPPARS-
Mflv_2856|M.gilvum_PYR-GCK DLGGDAAPITVRQHSVAPPAVDAKVRIHVTG---AAVVFGDRPRDGR---
Mb2419c|M.bovis_AF2122/97 TRG-DAEALALREGDTVYVRATRVPPIAGG---VSGVDDAGVERVKVTST
Rv2397c|M.tuberculosis_H37Rv TRG-DAEALALREGDTVYVRATRVPPIAGG---VSGVDDAGVERVKVTST
MAV_1785|M.avium_104 TRG-DAEALALREGDTVYVRATRVPPITAGATTVPAVSRDGADEATLTSA
MMAR_3715|M.marinum_M TRG-DAEALALQEGDTVYVRATRVPPIAGA---APAVPDDGAGSTPSAAQ
MUL_3658|M.ulcerans_Agy99 TRG-DAEALALQEGDTVYVRATRVPPIAGA---APAVPDDGAGSTPRAAQ
MAB_1655|M.abscessus_ATCC_1997 TRG-DAEALGLREGDPVYVRVTRVP--------------DVGELAEVAS-
MLBr_01424|M.leprae_Br4923 SKVTDQLVVARVDGRNPPAKGSRVRLYTEPG----NVHFFGVDGHRIS--
* :
MSMEG_3633|M.smegmatis_MC2_155 ------
Mvan_3091|M.vanbaalenii_PYR-1 GEAGS-
TH_3667|M.thermoresistible__bu ------
Mflv_2856|M.gilvum_PYR-GCK ------
Mb2419c|M.bovis_AF2122/97 ------
Rv2397c|M.tuberculosis_H37Rv ------
MAV_1785|M.avium_104 ------
MMAR_3715|M.marinum_M IGVGGQ
MUL_3658|M.ulcerans_Agy99 IGVGGQ
MAB_1655|M.abscessus_ATCC_1997 ------
MLBr_01424|M.leprae_Br4923 ------