For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAAEPTPDEIAALLQVEHLLDQRWPETKIEPGTGRISALLEMLGSPQQGYPSIHIGGTNGKTSVARMVD ALLTAFSRRTGRTTSPHLQSAVERISIDGVPISPAAYVDTYREIEPFVELVDQQSESAGGPKMSKFEVVT AMAFAAFADAPVDVAVVEVGLGGRWDATNVVNAPVAVITPIGVDHVDYLGGTIAEIAGEKAGIITRQADD LVPTDTVAVLARQVPEAMEVLLAEAVRADAAVAREDSEFAILGRQVAIGGQLLELQGLGGVYTDIFLPLH GEHQAHNAVVALAAVEAFFGAGTDRQLDVDTVRAGFAAVTSPGRLERMRNAPTVFIDAAHNPAGAAALSE ALRDEFDFRYLVGVVSVMGDKDVDGILAALEPALDQIVVTHNGSPRALEAAALAVKAEEIFGPERVITAA TLPDAIETATAVVEDSGSDGDAAGMSGAGIVITGSVVTAGAARTLFGKDPA
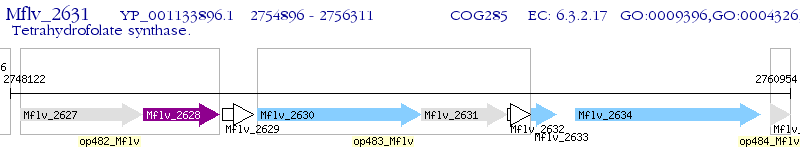
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2631 | - | - | 100% (471) | FolC bifunctional protein |
| M. gilvum PYR-GCK | Mflv_2983 | murE | 9e-07 | 25.07% (375) | UDP-N-acetylmuramoylalanyl-D-glutamate--2,6-diaminopimelate |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2474c | folC | 0.0 | 77.45% (470) | folylpolyglutamate synthase protein FolC |
| M. tuberculosis H37Rv | Rv2447c | folC | 0.0 | 77.45% (470) | folylpolyglutamate synthase protein FolC |
| M. leprae Br4923 | MLBr_01471 | folC | 0.0 | 71.34% (471) | folylpolyglutamate synthase |
| M. abscessus ATCC 19977 | MAB_1604 | - | 0.0 | 75.00% (472) | folylpolyglutamate synthase FolC |
| M. marinum M | MMAR_3771 | folC | 0.0 | 79.83% (471) | folylpolyglutamate synthase protein FolC |
| M. avium 104 | MAV_1725 | - | 0.0 | 78.25% (469) | FolC protein |
| M. smegmatis MC2 155 | MSMEG_4232 | murE | 2e-06 | 21.63% (416) | UDP-N-acetylmuramoylalanyl-D-glutamate--2,6-diaminopimelate |
| M. thermoresistible (build 8) | TH_0610 | folC | 0.0 | 78.27% (474) | PROBABLE FOLYLPOLYGLUTAMATE SYNTHASE PROTEIN FOLC |
| M. ulcerans Agy99 | MUL_3718 | folC | 0.0 | 79.62% (471) | folylpolyglutamate synthase protein FolC |
| M. vanbaalenii PYR-1 | Mvan_3950 | - | 0.0 | 90.66% (471) | FolC bifunctional protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2631|M.gilvum_PYR-GCK ---------------------------MSAAEPTPDEIAALLQVEHLLDQ
Mvan_3950|M.vanbaalenii_PYR-1 ---------------------------MSSFEPKPDEIAALLQVEHLLDQ
MMAR_3771|M.marinum_M -------------MSFDPPDW-TGLAQGSDAEPTPDELASLLQVEHLLDQ
MUL_3718|M.ulcerans_Agy99 -------------MSFDPPDW-TGLAQGSDAEPTPDELASLLQVEHLLDQ
MLBr_01471|M.leprae_Br4923 -------------MTAELPDWGSGRLETNVAALTPDKVASLLQVEHLLDQ
Mb2474c|M.bovis_AF2122/97 --------------MNSTNSGPPDSGSATGVVPTPDEIASLLQVEHLLDQ
Rv2447c|M.tuberculosis_H37Rv --------------MNSTNSGPPDSGSATGVVPTPDEIASLLQVEHLLDQ
MAV_1725|M.avium_104 --------------MTEPPDWAPEH-DVPDPVPTPDEIAALLQVEHLLDQ
TH_0610|M.thermoresistible__bu -------------------------MEMTQPAPTPDEIAALLQVEHLLDQ
MAB_1604|M.abscessus_ATCC_1997 ------------------------MSEPLSSEPSPDEIAALLQIEYLLDQ
MSMEG_4232|M.smegmatis_MC2_155 MAMKLRPSRPVGHHLAPLAAAVGAVASENPAGSSSDLAAVHVTGVTLRGQ
..* * : * .*
Mflv_2631|M.gilvum_PYR-GCK RWPETKIEPGTGRISALLEMLGSPQQGYPSIHIGG-TNGKTSVARMVDAL
Mvan_3950|M.vanbaalenii_PYR-1 RWPETKIEPSTARISALMELLGSPQLGYPSIHIGG-TNGKTSVARMVDSL
MMAR_3771|M.marinum_M RWPETRIEPSLTRISALMDLLGSPQLSYPSIHIGG-TNGKTSVARMVDAL
MUL_3718|M.ulcerans_Agy99 RWPETRIEPSLTRISALMDLLGSPQLSYPSIHIGG-TNGKTSVARMFDAL
MLBr_01471|M.leprae_Br4923 RWPETCIEPSLTRISALMDVLGSPQLSYPSIHVAG-TNGKTSVTRMVDAL
Mb2474c|M.bovis_AF2122/97 RWPETRIDPSLTRISALMDLLGSPQRSYPSIHIAG-TNGKTSVARMVDAL
Rv2447c|M.tuberculosis_H37Rv RWPETRIDPSLTRISALMDLLGSPQRSYPSIHIAG-TNGKTSVARMVDAL
MAV_1725|M.avium_104 RWPETKIEPSLTRISALLDLLGSPQRAYPSIHVAG-TNGKTSVARMIDAL
TH_0610|M.thermoresistible__bu RWPETKLDPSTARIAALMEMLGSPQRGYPTIHIAG-TNGKTSVARMVDAL
MAB_1604|M.abscessus_ATCC_1997 RWPETKIEPSTDRIVALMELLGSPQRAYPCIHVAG-TNGKTSVTRMIDAL
MSMEG_4232|M.smegmatis_MC2_155 DAQPGDLFAALPGASAHGARFAADAVAAGAVAVLTDPSGADLLAGTLDVP
: .. * :.: . : : ..* :: .*
Mflv_2631|M.gilvum_PYR-GCK LTAFSRR---TGRTTSPHLQSAVERISIDGVPISPAAYVDTY------RE
Mvan_3950|M.vanbaalenii_PYR-1 LTALHRR---TGRTISPHLQSAVERISIDGRPISPAAYVDTY------RE
MMAR_3771|M.marinum_M ITALHQR---TGRTTSPHLQSAVERISIDGKPITPAQYVQTY------RE
MUL_3718|M.ulcerans_Agy99 ITALHQR---TGRTTSPHLQSAVERISIDGKPITPAQYVQTY------RE
MLBr_01471|M.leprae_Br4923 LTALHQR---TGRFTSPHLQSVVERIAIDSKPISPAQYVATY------RE
Mb2474c|M.bovis_AF2122/97 VTALHRR---TGRTTSPHLQSPVERISIDGKPISPAQYVATY------RE
Rv2447c|M.tuberculosis_H37Rv VTALHRR---TGRTTSPHLQSPVERISIDGKPISPAQYVATY------RE
MAV_1725|M.avium_104 VTALHRR---TGRTTSPHLQSAVERISIDAKPISPARYVDTY------RE
TH_0610|M.thermoresistible__bu LRALHRR---TGRTTSPHLQSAVERIAIDGEPISPARYVEVY------RE
MAB_1604|M.abscessus_ATCC_1997 LTALHRR---TGRTTSPHLQSAVERIAIDGQPISPAKYVEIY------SE
MSMEG_4232|M.smegmatis_MC2_155 VLVHPAPRSVLGEAAAIVYGRPAERLRVVGVTGTSGKTTTTYLAEAGLRA
: . *. : .**: : . . :.. . *
Mflv_2631|M.gilvum_PYR-GCK IEPFVELVDQQSESAGG----PKMSKFEVVTAMAFAAFADAP-VDVAVVE
Mvan_3950|M.vanbaalenii_PYR-1 IEPFVELVDQQSQAAGG----PRMSKFEVVTAMAFAAFADAP-VDVAVVE
MMAR_3771|M.marinum_M IEPFVQMIDQQSQAAGG----PAMSKFEVLTAMAFAAFADAP-VDVAVVE
MUL_3718|M.ulcerans_Agy99 IEPFVQMIDQQSQAAGG----PAMSKFEVLTAMAFAAFADAP-VDVAVVE
MLBr_01471|M.leprae_Br4923 VEPFVQMIDAQSQSSGG----PALSKFEVVTAMAFAAFANAP-VDIAVVE
Mb2474c|M.bovis_AF2122/97 IEPLVALIDQQSQASAGKGG-PAMSKFEVLTAMAFAAFADAP-VDVAVVE
Rv2447c|M.tuberculosis_H37Rv IEPLVALIDQQSQASAGKGG-PAMSKFEVLTAMAFAAFADAP-VDVAVVE
MAV_1725|M.avium_104 IEPFVQMVDQQSQAAGG----PAMSKFEVLTAMAFAAFADAP-VDVAVVE
TH_0610|M.thermoresistible__bu IEPFVELVDKQSQAAGG----PALSKFEVVTAMAFAAFADAP-VDVAVVE
MAB_1604|M.abscessus_ATCC_1997 IEPFVELVDKQSQEQGG----PAMSKFEVLVAMAFVAFADAP-VDIAVVE
MSMEG_4232|M.smegmatis_MC2_155 AGRVAGLIGTVGVRIDGTDLPSALTTPEAPALQALLALMVERGVDTVVME
.. ::. . * . ::. *. . *: *: ** .*:*
Mflv_2631|M.gilvum_PYR-GCK VG-LGGRWDATNVVNAPVAVITPIGVDHVDYLGGTIAEIAGEKAGIITR-
Mvan_3950|M.vanbaalenii_PYR-1 VG-LGGRWDATNVINAPVAVVTPIGIDHADYLGDTIAEIAGEKAGIITR-
MMAR_3771|M.marinum_M VG-LGGRWDATNVINAPVAVITPISIDHVDYLGDDIASIAGEKAGIIHK-
MUL_3718|M.ulcerans_Agy99 VG-LGGRWDATNVINAPVAVITPISIDHVDYLGDDIASIAGEKAGIIHK-
MLBr_01471|M.leprae_Br4923 VG-LGGRWDATNVINASVAVITPVSIDHVEYLGNDVAAIAREKAGIITK-
Mb2474c|M.bovis_AF2122/97 VG-MGGRWDATNVINAPVAVITPISIDHVDYLGADIAGIAGEKAGIITR-
Rv2447c|M.tuberculosis_H37Rv VG-MGGRWDATNVINAPVAVITPISIDHVDYLGADIAGIAGEKAGIITR-
MAV_1725|M.avium_104 VG-MGGRWDATNVVNAPVAVITPISIDHVDYLGHDLAGIAGEKAGIITR-
TH_0610|M.thermoresistible__bu VG-MGGRWDATNIVNAPVAVITPIGVDHTDYLGDTVAEIAAEKAGIITRQ
MAB_1604|M.abscessus_ATCC_1997 VG-LGGRWDATNIIDAPVAVVTPIGIDHVEFLGPDLASIANEKAGIIKR-
MSMEG_4232|M.smegmatis_MC2_155 VSSHALAQGRVDGIGFAVGGFTNLSRDHLDFH-PTMADYFEAKARLFDP-
*. . . .: :. .*. .* :. ** :: :* ** ::
Mflv_2631|M.gilvum_PYR-GCK ----QADDLVPTDTVAVLARQVPEAMEVLLAEAVRADAAVAREDSEFAIL
Mvan_3950|M.vanbaalenii_PYR-1 ----QDDDLVPTDTVAVLARQVPEAMEVLLAQAVRCDAAVAREDSEFTVL
MMAR_3771|M.marinum_M ----AAD--GSPDTVAVIGRQLPEAMEVLLAQAVRADAAVAREDSEFAVL
MUL_3718|M.ulcerans_Agy99 ----AAD--GSPDTVAVIGRQLPEAMEVLLAQAVRADAAVAREDSEFAVL
MLBr_01471|M.leprae_Br4923 ----APE--GAPDTVVVIGRQVPAAMGVLLAQSAHAGARVVRQDSEFAVL
Mb2474c|M.bovis_AF2122/97 ----APD--GSPDTVAVIGRQVPKVMEVLLAESVRADASVAREDSEFAVL
Rv2447c|M.tuberculosis_H37Rv ----APD--GSPDTVAVIGRQVPKVMEVLLAESVRADASVAREDSEFAVL
MAV_1725|M.avium_104 ----APA--GAPDTVAVIAQQPPEVMEVLLAQCVQADAAVARAGSEFAVL
TH_0610|M.thermoresistible__bu DPGLVPEG-TDPSTVAVIGRQPPEAMEAILREAVRADAVVARADAEFAVI
MAB_1604|M.abscessus_ATCC_1997 HKLDAMTGEAGADTVAVIAQQQPEAMEVLLRQTVEADAAVAREGSEFAVL
MSMEG_4232|M.smegmatis_MC2_155 -----QSPNHAAAAVICVDDQYGVQMAARAETPLTVSTSGAAADWRVEKI
. :* : * * . .: . . .. :
Mflv_2631|M.gilvum_PYR-GCK GRQVAIGGQLLELQGLGGVYTDIFLPLHGEHQAHNAVVALAAVEAFFGAG
Mvan_3950|M.vanbaalenii_PYR-1 GRQVAIGGQLLELQGLGGVYPDIFLPLHGEHQAHNAVLALAAVEAFFGAG
MMAR_3771|M.marinum_M GRQVAVGGQLLQLQGLGGVYSDIYLPLHGEHQAHNAVVALAAVEAFFGAG
MUL_3718|M.ulcerans_Agy99 GRQVAVGGQLLQLQGLGGVYSDIYLPLHGEHQAHNAVVALAAVEAFFGAG
MLBr_01471|M.leprae_Br4923 DRQVAIGGQVLQLQGLGGVYNDIYLPLHGEHQAHNAVLVLAAVEAFYGAG
Mb2474c|M.bovis_AF2122/97 RRQIAVGGQVLQLQGLGGVYSDIYLPLHGEHQAHNAVLALASVEAFFGAG
Rv2447c|M.tuberculosis_H37Rv RRQIAVGGQVLQLQGLGGVYSDIYLPLHGEHQAHNAVLALASVEAFFGAG
MAV_1725|M.avium_104 GRQVAIGGQLLQLQGLGGVYSDVYLPLHGEHQAHNAALALAAVEAFFGAG
TH_0610|M.thermoresistible__bu GRQVAVGGQMLELRGLGGIYSEIFLPLHGEHQAHNAALALAAVEAFFGAG
MAB_1604|M.abscessus_ATCC_1997 SRQVAVGGQLLELQGLGGVYPEIFLPLHGEHQAHNAAVALAAVEAFFGAG
MSMEG_4232|M.smegmatis_MC2_155 T-TAGPGAQEFVLVDPGGMHHQLGVGLPGRYNVANAALAVALLDAVG---
. *.* : * . **:: :: : * *.::. **.:.:* ::*.
Mflv_2631|M.gilvum_PYR-GCK TDRQLDVDTVRAGFAAVTSPGRLERMRNAP--TVFIDAAHNPAGAAALSE
Mvan_3950|M.vanbaalenii_PYR-1 AQRQLDVDAVRAGFAAVTSPGRLERLRSAP--TVFIDAAHNPAGAAALVE
MMAR_3771|M.marinum_M AQRQLDVDAVRAGFAAVTSPGRLERLRSSP--TVFIDAAHNPAGAAALAQ
MUL_3718|M.ulcerans_Agy99 AQRQLDVDAVRAGFAAVTSPGRLERLRSSP--TVFIDAAHNPAGAAALAQ
MLBr_01471|M.leprae_Br4923 VQRQLDVEAVRSGFAAVISPGRLERMRSAP--TVFIDAAHNPAGANALAR
Mb2474c|M.bovis_AF2122/97 AQRQLDGDAVRAGFAAVTSPGRLERMRSAP--TVFIDAAHNPAGASALAQ
Rv2447c|M.tuberculosis_H37Rv AQRQLDGDAVRAGFAAVTSPGRLERMRSAP--TVFIDAAHNPAGASALAQ
MAV_1725|M.avium_104 AQRQLDVDAVRAGFAAVTSPGRLERMRSAP--TVFIDAAHNPAGAAALAR
TH_0610|M.thermoresistible__bu AQRQLDVDAVREGFASVINPGRMERVRSAP--TVFVDAAHNPAGAAALAE
MAB_1604|M.abscessus_ATCC_1997 ADRQLDIEAIRSGFASVIIPGRLERVRSAP--AVFLDAAHNPHGAAALAE
MSMEG_4232|M.smegmatis_MC2_155 ----VSPEQAAPGLRTATVPGRLESIDLGQDFLALVDYAHKPGALQAVLE
:. : *: :. ***:* : . .::* **:* . *: .
Mflv_2631|M.gilvum_PYR-GCK ALRDEFDFRYLVGVVSVMGDKDVDG--ILAALEPALDQIVVTHNGSPR--
Mvan_3950|M.vanbaalenii_PYR-1 ALRDEFDFRYLVGVVSVMGDKDVDG--ILAALETAFDQIVVTHNGSPR--
MMAR_3771|M.marinum_M SLVAEFDFRSLVGVIAVMADKDVNG--ILAALEPAFDSIVVTHNGSPR--
MUL_3718|M.ulcerans_Agy99 SLVAEFDFRSLVGVVAVMADKDVNG--ILAALEPAFDSIVVTHNGSPR--
MLBr_01471|M.leprae_Br4923 TLSDEFNFRTLVGVLSVLANKDVDG--ILAALELVLDYIVVTDNGSPR--
Mb2474c|M.bovis_AF2122/97 TLAHEFDFRFLVGVLSVLGDKDVDG--ILAALEPVFDSVVVTHNGSPR--
Rv2447c|M.tuberculosis_H37Rv TLAHEFDFRFLVGVLSVLGDKDVDG--ILAALEPVFDSVVVTHNGSPR--
MAV_1725|M.avium_104 TLADEFDFRFLVGVLSVLADKDVDG--ILAALQPVFHSVVVTHNGSPR--
TH_0610|M.thermoresistible__bu TLQSEFDFRFLVGVVSVLAGKDVDG--ILAALEPVFDQIIITHNGSPR--
MAB_1604|M.abscessus_ATCC_1997 TLQSEFDFRRLVGVISVMGDKDVAG--ILGALEPAFDEIVVTHNGSPR--
MSMEG_4232|M.smegmatis_MC2_155 TLR-AAEPRRIAVVFGAGGDRDPGKREPMGRVTAELADLVVVTDDNPRSE
:* : * :. *... ..:* :. : : :::. :..**
Mflv_2631|M.gilvum_PYR-GCK --ALEAAALAVKAEEIFGPERVITAATLPDAIETATAVVEDS-----GSD
Mvan_3950|M.vanbaalenii_PYR-1 --ALEVVALAVKAEEIFGPERVVTASTLPDAIETATALVEES-----GSA
MMAR_3771|M.marinum_M --ALDVASLALAAEQYFGPDRVITAENLRDAVDAATALVDES-----AID
MUL_3718|M.ulcerans_Agy99 --ALDVASLALAAEQYFGPDRVITAENLRDAVDAATALVDES-----AID
MLBr_01471|M.leprae_Br4923 --ALDVESLALVARERFGPDRVISAANLRDAIDAATALVDDA-----AAN
Mb2474c|M.bovis_AF2122/97 --ALDVEALALAAGERFGPDRVRTAENLRDAIDVATSLVDDA-----AAD
Rv2447c|M.tuberculosis_H37Rv --ALDVEALALAAGERFGPDRVRTAENLRDAIDVATSLVDDA-----AAD
MAV_1725|M.avium_104 --ALDVESLALAAREQFGPDRVSTAENLSDAIDVATALVDDA-----VRD
TH_0610|M.thermoresistible__bu --ALEIPVLAARAEERFGEERVLTADTLADAIETATALVEDA-----GME
MAB_1604|M.abscessus_ATCC_1997 --AMETDALAGIALEIFGEDRVVVAPTLLDAVETATAMVEEA-----AED
MSMEG_4232|M.smegmatis_MC2_155 DPATIRSAILAGARAVASDAEIVEIGDRRAAIDHAVAWAQPGDIVLIAGK
* : * . .: *:: *.: .: .
Mflv_2631|M.gilvum_PYR-GCK GDAAG----MSGAGIVITGSVVTAGAARTLFGKDPA---
Mvan_3950|M.vanbaalenii_PYR-1 GDAAG----MGGAGIVITGSVVTAGAARTLFGKDPA---
MMAR_3771|M.marinum_M WEAQG--GSLSGTGIVITGSVVTAGAARTLFGRDPQ---
MUL_3718|M.ulcerans_Agy99 WEAQG--GSLSGTGIVITGSVVTAGAAHTLFGRDPQ---
MLBr_01471|M.leprae_Br4923 EEIEG--LGVSGRGIVITGSVITAGIARTLFGRDPQ---
Mb2474c|M.bovis_AF2122/97 PDVAG--DAFSRTGIVITGSVVTAGAARTLFGRDPQ---
Rv2447c|M.tuberculosis_H37Rv PDVAG--DAFSRTGIVITGSVVTAGAARTLFGRDPQ---
MAV_1725|M.avium_104 GLVDGQAEDFAGTGIVITGSVVTAGAARTLFGRDPQ---
TH_0610|M.thermoresistible__bu G--EG----FSGSGIVFTGSVVTAGAARSLFGKEPQ---
MAB_1604|M.abscessus_ATCC_1997 GGAEG----FSGTGIVITGSVVTAGAARTLFGKDPQ---
MSMEG_4232|M.smegmatis_MC2_155 GHESG---QTVGDHTRPFDDRDELAAALEASGKTRDRSS
* .. . * *: