For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAAPDPNPVSPTPFHDLDEFLSLPRVSGLAVSRDGARVVTTVATLNDTRTEFVSALWELDPVGREPARR ITRGVTGESAPEFTADGDLLFLAARPTADDDKPPKSLWCLPVAGGEAYEVCQMPGGVDGVASAGGATVVV SSMLPSAVDIDADERLRTARKDNKVSALLHTGYPVRAWDHDIGPARTHLFDADTKRDVTPDPGDALRDSH FDVSPDGGFLVASWNVPAPGAALRSTLVRIDVNTGDRAVIVDDPESDAGFPAIAPDGTSVAYLRETLSTP HHAPRITLALLHFGEEPRELTAHWDRWPTSVTWSADGSTLLVTADDDGRGPIFAVDPQTGAVRTLVSDDY TYTDVRAAPGGVIFALRSSYAAPQHPVRIDPDGTITVLDCVPLPSLPGTLTEIEATAADGQRVRSWLVLP EGDHPAPLLLWIHGGPLASWNAWHWRWNPWLMAAGGYAVLLPDPALSTGYGQEFIERGWGAWGFAPYTDL MAATDAACAHPRIDETRTAVMGGSFGGYMANWIAGHTDRFAAIVTHAGLWALDQFGATTDGSYWWSREMT PKMASANSPHLFVSDIRTPMLVIHGDKDYRVPIGEALRLWYDLLSRSGLPAADSGPEAGTTQHRFLYFPS ENHWILSPQHAKLWYQVVLGFLNLHVLGQDADVPELLG
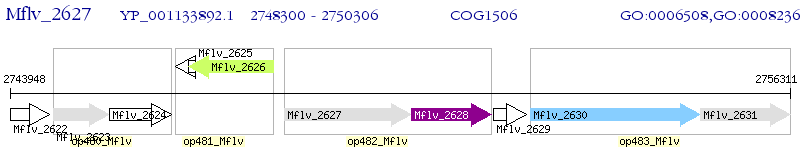
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2627 | - | - | 100% (668) | peptidase S9 prolyl oligopeptidase |
| M. gilvum PYR-GCK | Mflv_4393 | - | 2e-19 | 30.42% (286) | peptidase S9 prolyl oligopeptidase |
| M. gilvum PYR-GCK | Mflv_2548 | - | 1e-13 | 26.83% (246) | peptidase S9 prolyl oligopeptidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4725 | - | 5e-23 | 28.79% (448) | peptidase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4633 | - | 0.0 | 68.67% (667) | peptidase S9, prolyl oligopeptidase |
| M. thermoresistible (build 8) | TH_4519 | - | 6e-15 | 24.52% (367) | PUTATIVE peptidase S9, prolyl oligopeptidase active site domain |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3954 | - | 0.0 | 75.51% (686) | peptidase S9 prolyl oligopeptidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2627|M.gilvum_PYR-GCK ------------MTAAPD-PNPVSP--TPFHDLDEFLSLPRVSGLAVSRD
Mvan_3954|M.vanbaalenii_PYR-1 MKSCPAMCYEFRMSSAPEQPTPATPGTTAFHDLDEFLALPRVAGLAVSPD
MSMEG_4633|M.smegmatis_MC2_155 ------------MN------------STPFHDLDAYLDLPRVSGLAVSPD
MAB_4725|M.abscessus_ATCC_1997 ------------------------MGLPKLITVEEFFEPPVRTSASISPD
TH_4519|M.thermoresistible__bu ----------------------------------VAEQVRATYGASLSPD
. ::* *
Mflv_2627|M.gilvum_PYR-GCK GARVVTTVATLNDTRTEFVSALWELDPVGREPARRITRGVTGESAPEFTA
Mvan_3954|M.vanbaalenii_PYR-1 GTRVVTTVSRLDDKRTEFVSALWELDPSGQQPARRLTFGAKGESAPEFTA
MSMEG_4633|M.smegmatis_MC2_155 GTRVVTTVSTLNAKRTEFVTALWELDPAGQQPARRLTRGLKGESAPVFTA
MAB_4725|M.abscessus_ATCC_1997 GTMIAYLAPRKNR------LNVWVQG-IGSTEARCVTHDETRSVTGFFWV
TH_4519|M.thermoresistible__bu ATAFAHLVDDGGFPR----AVQRFLRGWRASSSRDVELPVVGPVRRVLHS
.: .. . . :* : :
Mflv_2627|M.gilvum_PYR-GCK DGDLLFLAARPTADDDKPP-KSLWCLPVAGGEAYEVCQMPGGVDGVASA-
Mvan_3954|M.vanbaalenii_PYR-1 DGDLLFLAARPTGDDDEPTRKALWRLPAAGGEAHEVATLPGGVDRVTSAR
MSMEG_4633|M.smegmatis_MC2_155 DGDVLFLAVRPTEDDEKPP-AALWRLPAHGGEAHEMLSLPGGVSAAASAR
MAB_4725|M.abscessus_ATCC_1997 DDPRWLLYVQDSGGDENWH-LYRVDLSTPDATAVDLTPFPGARLLGLERP
TH_4519|M.thermoresistible__bu PDGGWLACEVAPDGSDRSQ-IWVVTTDPDDRGAHRIDSWPPDAPEGAAEL
. : . ..:. . * : *
Mflv_2627|M.gilvum_PYR-GCK -GGATVVVSSMLPSAVDIDADERLRTARKDNKVSALLHTGYPVRAWDHD-
Mvan_3954|M.vanbaalenii_PYR-1 EAATTLITSAMLASATDVDDDKRLRAARKDNKVSAVLHAGYPVRYWDHD-
MSMEG_4633|M.smegmatis_MC2_155 TADVTVVAAPLLPSAADVEADRSLRKARKDNKVSAILHTGYPVRQWDHD-
MAB_4725|M.abscessus_ATCC_1997 YGRSGKVYIQLNHRRIDQIDLYELDIASGELTMLAENPGGVMAWFCGPNH
TH_4519|M.thermoresistible__bu IAWDGNRIAAILTGQDGVGSSCLIDPADGSTVVLDRRSGGRLVDAWAGAA
. : . : * . : * .
Mflv_2627|M.gilvum_PYR-GCK --IGPARTHLFDADTKR--DVTPDPGDALRDSHFDVSPDGGFLVASWNVP
Mvan_3954|M.vanbaalenii_PYR-1 --LGPEQPHLFDITEERPRDLTPTPGQALREAHFDLSRDGAFLVTTWQGP
MSMEG_4633|M.smegmatis_MC2_155 --LGPDLPHLFDADGAR--NLTERPGVALRESTFDVSPDGAFVVATWHVP
MAB_4725|M.abscessus_ATCC_1997 EVFAQSLTPGGDIEFSRYESGALRPLCVYDGADYPVGMNPTQMTPDGSGM
TH_4519|M.thermoresistible__bu LIRVGPRGYRELIMLHGNTEMALLPSDPGSTTDFGVILDDHSARRLRRGP
. : * : : : :
Mflv_2627|M.gilvum_PYR-GCK APG---AALRSTLVRIDVNTGDRAVIVDDPESDAGFPAIAPDGTSVAYLR
Mvan_3954|M.vanbaalenii_PYR-1 APG---AALRTTLVHVDVATGRRTTIVDDRDADLGNPAVSPDGQSVAYTR
MSMEG_4633|M.smegmatis_MC2_155 APG---AATRSTLVRIDRETGARTTIAEDPQADLEHPVIAPDGGALAFTR
MAB_4725|M.abscessus_ATCC_1997 WLGSSRDTDRTRLVRLDLATGAESTIDSHPAYDLAAALGLPP----TLIR
TH_4519|M.thermoresistible__bu DGE-----LTKLYQPAKRYPRESTEGYVRALIRSDNGAGYARLLEVTTTA
. . . : . :
Mflv_2627|M.gilvum_PYR-GCK ETLSTPHHAPRITLALLHFGEEP------RELTAHWDRWPTSVTWSADGS
Mvan_3954|M.vanbaalenii_PYR-1 ETISTPRKAPRITLCLLRFGEEP------VELTTGWDRWPTSVTWSADGS
MSMEG_4633|M.smegmatis_MC2_155 ETHSTPTEPPRITLCCMRFGNDDEQLGEWAEVATEWDRWPSSLAWSADAS
MAB_4725|M.abscessus_ATCC_1997 HRKSGELLAARYLGARQEI----------RVLDPDFGPVLEKLQELSDGD
TH_4519|M.thermoresistible__bu DGVAYQVVAERPGHDLDEF-----------VVSDDLSTVALLWNIDGASE
. : . * .: : . . ..
Mflv_2627|M.gilvum_PYR-GCK TLLVTADDDGRGPIFAVDPQTGAVRTLVSDDYTYTDVRAAPGGVIFALRS
Mvan_3954|M.vanbaalenii_PYR-1 SVIITADDHGRGPIFAVDPDTGAVRTLVADDFTYSDVRPAPGGVLFALRS
MSMEG_4633|M.smegmatis_MC2_155 ALIVTADDNGRGPVFSVHPDTGTVTRLTDDDHTYTDVCPAPGGIIYALRS
MAB_4725|M.abscessus_ATCC_1997 LAEVSSDEDGRRWVVSFTHDR-------------------DPGVTYFYDH
TH_4519|M.thermoresistible__bu LQILQYADNTLSEPIPLPGMVAG------------ELSISAGGSMVAMTV
: :. ... *
Mflv_2627|M.gilvum_PYR-GCK SYAAPQHPVRIDPDGTITVLDCVPLPSL---PGTLTEIEATAADGQRVRS
Mvan_3954|M.vanbaalenii_PYR-1 SYAAPQHPVRIDADGTVTVLDCVALPDL---PGTLTEVTATTADGRTVRS
MSMEG_4633|M.smegmatis_MC2_155 SYAVPPHPVRIDPDGTVTELPCFDAPAL---PGSLTEITATAEDGTPVRS
MAB_4725|M.abscessus_ATCC_1997 ATGESHQLFRPYP---------HLDPAD---LAPVVPVTITARDGLTLNS
TH_4519|M.thermoresistible__bu ESPWLPPTVELVDPRTRQWELIDREPSTGPVTGTPTLERLTARDGLELTG
.. * .. . *: ** : .
Mflv_2627|M.gilvum_PYR-GCK WLVLP-EGDHP----------APLLLWIHGGPLASWNAWHWRWNPWLMAA
Mvan_3954|M.vanbaalenii_PYR-1 WMVLP-HGEAAGSYSSRERSSAPLLLWIHGGPLASWNAWHWRWNPWVMAA
MSMEG_4633|M.smegmatis_MC2_155 WLTLP-ESTDP----------APLVLWVHGGPLGSWNTWHWRWNPWLLTA
MAB_4725|M.abscessus_ATCC_1997 YLTLP-VGTEP--------EGLPLVLVVHGGPWH-RDSWGFDPTVQLLAN
TH_4519|M.thermoresistible__bu WLYRPPTGVQP----------VGAMIFLHGGPEGQSRPGYNEYFPPLLDR
:: * . . :: :**** . ::
Mflv_2627|M.gilvum_PYR-GCK GGYAVLLPDPALSTGYGQEFIERGWGAWGFAPYTDLMAATDAACAHPRID
Mvan_3954|M.vanbaalenii_PYR-1 RGYAVLLPDPALSTGYGQEFIQRGWGSWGFAPYTDLMAATDAACAHPRVD
MSMEG_4633|M.smegmatis_MC2_155 HGYAVLMPDPALSTGYGQDFIARGWGAWGEAPYTDLMAATDAACEHPRVD
MAB_4725|M.abscessus_ATCC_1997 RGYAVLQVNFRGSTGYGKAFTKAAIGEFAGKMHDDLIDAVDWAVEQGYAD
TH_4519|M.thermoresistible__bu -GITVFTPNVRGSGGFGRSFVHADDKENRFRAIDDVADCVHYLVRNGLAP
* :*: : * *:*: * *: ... :
Mflv_2627|M.gilvum_PYR-GCK ETRTAVMGGSFGGYMANWIAGHT-DRFAAIVTHAGLWALDQFGATTD---
Mvan_3954|M.vanbaalenii_PYR-1 ETRTAAMGGSFGGYMANWIAGHT-DRFAAIVTHASLWALDQFGATTD---
MSMEG_4633|M.smegmatis_MC2_155 ASRTAAMGGSFGGYMANWVAGHT-DRFDAIVTHASLWALDQFAPTTD---
MAB_4725|M.abscessus_ATCC_1997 PGRIAIFGGSYGGYSALVGVTFTPDVFAAAVDYVGISNLANFMRTLPPFV
TH_4519|M.thermoresistible__bu ADRIACAGWSYGGYLTQAALAFHPDLFAAGISVCGMSDLTTWYQTTEP--
* * * *:*** : . * * * : .: * : *
Mflv_2627|M.gilvum_PYR-GCK ----GSYWWSREMTPKMAS------ANSPHLFVSDIRTPMLVIHGDKDYR
Mvan_3954|M.vanbaalenii_PYR-1 ----GGYWWAREMTPEMAA------ANSPHLFVSQIATPMLVIHGDKDYR
MSMEG_4633|M.smegmatis_MC2_155 ----GAYWWAREMTPEMMQ------RNSPHRFVDQINTPMLVIHGDKDYR
MAB_4725|M.abscessus_ATCC_1997 RPNLANNWYRYVGDPAVPEQEADMLARSPISRVDRIRTPLLVAQGANDVR
TH_4519|M.thermoresistible__bu --WIAQSSYTEYGHPVADR--DLLERLSPLTHAAAVTAPLLLVHGASDTN
. : * ** . : :*:*: :* .* .
Mflv_2627|M.gilvum_PYR-GCK VPIGEALRLWYDLLSRSGLPAADSGPEAGTTQHRFLYFPSENHWILSPQH
Mvan_3954|M.vanbaalenii_PYR-1 VPIGEALRLWYELLSSSALPAADDG----STEHRFLYFPSENHWVLSPQH
MSMEG_4633|M.smegmatis_MC2_155 VPIGEALRLWYELLTESALPADDNG----DSPHRFLYYPAENHWVLTPQH
MAB_4725|M.abscessus_ATCC_1997 VVQAESDNIVAALRARG-------------VEVEYMVKADEGHGFLNPEN
TH_4519|M.thermoresistible__bu VPPSESIQMHTALERLG-------------RPVKLLMFDDDGHEIVRREN
* .*: .: * . . : :.* .: ::
Mflv_2627|M.gilvum_PYR-GCK AKLWYQVVLGFLNLHVLGQDADVPELLG
Mvan_3954|M.vanbaalenii_PYR-1 AKIWYQVVLAFLAHHVLGQDSELPEMLG
MSMEG_4633|M.smegmatis_MC2_155 AKLWYQVVTAFLDENLRNEPAQWPELLG
MAB_4725|M.abscessus_ATCC_1997 QIDLHRATERFLAQHVGGRQVS------
TH_4519|M.thermoresistible__bu RATLVEAMSHWLMSAFAERRREAS----
.. :* . . .