For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPEVEDALSTMASVNNEFFYWMSIALMMLIHAGFLAYEIGASRSKNVLATAMKNLLAFATIVASFFFVGW FLYNSMPSGFIEFNDAAKAALPWSDNMGPNTDDPASGIFWGAFALFAATTGSIMSGAVLERIRTSGFLVL TVFVGSVVWIIGASWGWHGAGWMLTELGFHDVGAAGCVHMIAGFAALGILINLGPRIGRFLPDGTAVTIR PHNLPLTMVGLFLIFTGFFGFLMGCVIYGPGGYATIYGSPTTLSAFAFNTLMGLAGGIIGAYLTSRGEPF WTISGGLAGVIAVAAGMDLYHPALTFVIALVGGGLIPFIGKLLDKMKIDDVVGAVAVHGGMGFYSLIMAG IFLAGYPNTDGNPSISFWGQLIGAVTFAALGFLPAYLLSLGLKKAGLLRIPPEVEEQGLDLSEVPATPYP EGIPVTEMRASSGTAVLVLGEEVK*
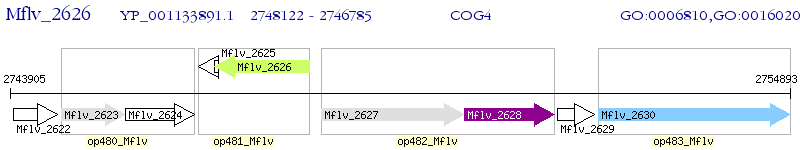
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2626 | - | - | 100% (445) | Rh family protein/ammonium transporter |
| M. gilvum PYR-GCK | Mflv_1301 | - | 5e-22 | 25.80% (314) | ammonium transporter |
| M. gilvum PYR-GCK | Mflv_4185 | - | 3e-16 | 26.20% (439) | ammonium transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2944c | amt | 4e-23 | 26.19% (443) | ammonium transporter |
| M. tuberculosis H37Rv | Rv2920c | amt | 4e-23 | 26.19% (443) | ammonium transporter |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0355 | - | 3e-23 | 26.43% (420) | ammonium transporter |
| M. marinum M | MMAR_0038 | amt_1 | 7e-32 | 26.82% (425) | ammonium-transport integral membrane protein, Amt_1 |
| M. avium 104 | MAV_3084 | amt | 2e-29 | 26.43% (420) | ammonium transporter |
| M. smegmatis MC2 155 | MSMEG_4635 | - | 0.0 | 83.61% (427) | ammonium transporter family protein |
| M. thermoresistible (build 8) | TH_4321 | - | 1e-20 | 25.67% (448) | PUTATIVE ammonium transporter |
| M. ulcerans Agy99 | MUL_0037 | amt_1 | 2e-31 | 26.82% (425) | ammonium-transport integral membrane protein, Amt_1 |
| M. vanbaalenii PYR-1 | Mvan_3955 | - | 0.0 | 90.32% (444) | Rh family protein/ammonium transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2944c|M.bovis_AF2122/97 -MDQFP--IMGVPDGGDTAWMLVSSALVLLMTPGLAFFYGGMVRSKSVLN
Rv2920c|M.tuberculosis_H37Rv -MDQFP--IMGVPDGGDTAWMLVSSALVLLMTPGLAFFYGGMVRSKSVLN
MAB_0355|M.abscessus_ATCC_1997 ---MFP--HMGVPDAANTAWVLTSAALVLLMTPALAFFYGGMVRAKSVLN
TH_4321|M.thermoresistible__bu VVLALPDESFAAFDSGDTAWVLISAALVLLMTPGLAFFYGGLSRQKSVLN
MMAR_0038|M.marinum_M ----------MDIDPAATAWLLASTALVLLMTPGLAIFYGGMVRTTGVLN
MUL_0037|M.ulcerans_Agy99 ----------MDIDPAATAWLLASTALVLLMTPGLAIFYGGMVRTTGVLN
MAV_3084|M.avium_104 ---------MHGIDPAATAWLLASTALVLLMTPGLAIFYGGMVRTTGVLN
Mflv_2626|M.gilvum_PYR-GCK MTPEVEDALSTMASVNNEFFYWMSIALMMLIHAGFLAYEIGASRSKNVLA
Mvan_3955|M.vanbaalenii_PYR-1 MTPEVEDAIASMTAVNNEFFYWMSIALMMLIHAGFLAYEIGASRSKNVLA
MSMEG_4635|M.smegmatis_MC2_155 MPPDVQDALDTMATINNEFYYWVSIALMFLIHAGFLAYEVGASRSKNVLA
: * **::*: ..: : * * ..**
Mb2944c|M.bovis_AF2122/97 MIMMSISAMGVVTVLWALYGYSIAFGDDVG------NIAGNPSQYWGLKG
Rv2920c|M.tuberculosis_H37Rv MIMMSISAMGVVTVLWALYGYSIAFGDDVG------NIAGNPSQYWGLKG
MAB_0355|M.abscessus_ATCC_1997 MMMMCFSAVAVVWLLWVIFGYSMAFGTDIGG-----GLLGDPFQFAGLQH
TH_4321|M.thermoresistible__bu MMMMSFGALGIVSVVYVLWGYSMSFSSGHTGQSDILGVFDNPFSLFGLSP
MMAR_0038|M.marinum_M MIMMSFISIPLVTVAWLLAGYSLAFSDGAA-----PGFPGGLVGGLTHAG
MUL_0037|M.ulcerans_Agy99 MIMMSFISIPLVTVAWLLAGYSLAFSDGAA-----PGLPGGLVGGLTHAG
MAV_3084|M.avium_104 MIMMSFISIPLVTVAWLLVGYTMAFSQ--------DGMSG-LVGNLRHFG
Mflv_2626|M.gilvum_PYR-GCK TAMKNLLAFATIVASFFFVGWFLYNSMPSG----FIEFNDAAKAALPWSD
Mvan_3955|M.vanbaalenii_PYR-1 TAMKNLLAFATIVASFFFVGWFLYNAMPTG----FIEFSDAAKAALPWSD
MSMEG_4635|M.smegmatis_MC2_155 TAMKNLLALATIIASFYFVGWFLYNAMPSG----FIEFNEAAKAALPWGD
* : :. : : : *: : . .
Mb2944c|M.bovis_AF2122/97 LIGVNAVAADPSTQTAAVNIPLAG---TLPATVFVAFQLMFAIITVALIS
Rv2920c|M.tuberculosis_H37Rv LIGVNAVAADPSTQTAAVNIPLAG---TLPATVFVAFQLMFAIITVALIS
MAB_0355|M.abscessus_ATCC_1997 LFEG---------DYKAPSVPLWGPT-NIPALVFVMFQAGFAMVTVALIA
TH_4321|M.thermoresistible__bu LLETR--------EIDGATLSVIGGFGTVPAIVWVGFQLTFAVITVALIS
MMAR_0038|M.marinum_M MRGIG-------------PATVHG---SVPELLFATFQLSFAIITAALVS
MUL_0037|M.ulcerans_Agy99 MRGIG-------------PATVHG---SVPELLFATFQLSFAIITAALVS
MAV_3084|M.avium_104 MLGIT-------------PDTTHG---AVPELLYATFQLTFAIITAALVS
Mflv_2626|M.gilvum_PYR-GCK NMGPN------------TDDPASG--------IFWGAFALFAATTGSIMS
Mvan_3955|M.vanbaalenii_PYR-1 NMGPN------------TADAASG--------IFWGAFALFAATTGSIMS
MSMEG_4635|M.smegmatis_MC2_155 NMGPN------------VQDSASG--------IFWGAFALFAATTGSIMS
. * :: ** * ::::
Mb2944c|M.bovis_AF2122/97 GAVADRLKFGAWLLFAGLWATFVYFPVAHWVF-------AFDGFAAEHGG
Rv2920c|M.tuberculosis_H37Rv GAVADRLKFGAWLLFAGLWATFVYFPVAHWVF-------AFDGFAAEHGG
MAB_0355|M.abscessus_ATCC_1997 GAVADRMRFLPWIAFTALWATLVYFPVAHWIF-------AVEGTTADKGG
TH_4321|M.thermoresistible__bu GALAERVRFGTWIVFAAVWVTLVYFPLAHMVWGGGLLSDAQNGFASWLFG
MMAR_0038|M.marinum_M GAIADRAKFAAWMVFVPVWSIAVYSVVAHWVWS--------------PSG
MUL_0037|M.ulcerans_Agy99 GAIADRAKFAAWMVFVPVWSIAVYSVVAHWVWA--------------PSG
MAV_3084|M.avium_104 GAIADRAKFAAWMVFVPVWTVAVYSVVAHWVWG--------------PGG
Mflv_2626|M.gilvum_PYR-GCK GAVLERIRTSGFLVLTVFVGSVVWIIGASWGWHG--------------AG
Mvan_3955|M.vanbaalenii_PYR-1 GAVLERIRTSGFLILTVFVGSVLWIIGASWGWHG--------------AG
MSMEG_4635|M.smegmatis_MC2_155 GAVLERIRTSGFLILTVLVGSVTWIIGAAWGWHG--------------AG
**: :* : :: :. . : * : *
Mb2944c|M.bovis_AF2122/97 WIANK----LHAIDFAGGTAVHINAGVAALMLAIVLGKRRG-----WPAT
Rv2920c|M.tuberculosis_H37Rv WIANK----LHAIDFAGGTAVHINAGVAALMLAIVLGKRRG-----WPAT
MAB_0355|M.abscessus_ATCC_1997 WLAN-----LGVLDFAGGTAVEINSGAAGLALAIVLGKRRG-----WPRE
TH_4321|M.thermoresistible__bu FNEDEGAANVAPIDFAGGTVVHINAGMAALMLALVVGRRLG-----FGRT
MMAR_0038|M.marinum_M WLAK-----MGVLDYAGGLVVEIVSGSSALALALVLGPRIG-----FKVD
MUL_0037|M.ulcerans_Agy99 WLAK-----MGVLDYAGGLVVEIVSGSSARALAPVLGPRIG-----FKVD
MAV_3084|M.avium_104 WLAR-----LGVLDYAGGLVVEIVSGSSALALALVLGPRIG-----FKKD
Mflv_2626|M.gilvum_PYR-GCK WMLTE----LGFHDVGAAGCVHMIAGFAALGILINLGPRIGRFLPDGTAV
Mvan_3955|M.vanbaalenii_PYR-1 WMLTK----LGFHDVGAAGCVHMIAGFATLGILINLGPRIGRFLPDGTAV
MSMEG_4635|M.smegmatis_MC2_155 WMLTK----LGFHDVGAAGCVHMIAGFATLGVLINLGPRIGRFLPDGTPV
: : * ... *.: :* : : :* * *
Mb2944c|M.bovis_AF2122/97 LFRPHNLPFVMLGAALLWFGWYGFNAGSATTANGVAGATFVTTTIATAAA
Rv2920c|M.tuberculosis_H37Rv LFRPHNLPFVMLGAALLWFGWYGFNAGSATTANGVAGATFVTTTIATAAA
MAB_0355|M.abscessus_ATCC_1997 AMRPHNLPAVMLGAGLLWFGWFGFNAGSALAADGTAALVIANTLGAGAIS
TH_4321|M.thermoresistible__bu AYRPHNIPWVMLGAALLWFGWFGFNVGSEGAADAVAGLVWVNTTAATAAA
MMAR_0038|M.marinum_M PMRPHNLPFVLLGVGLLWFGWFGFNAGSALAANGLASAIFLNTLVAGCLG
MUL_0037|M.ulcerans_Agy99 PMRPHNLPFVLLGVGLLWFGWFGFNAGSALAANGLASAIFLNTLVAGCLG
MAV_3084|M.avium_104 AMRPHNLPLLFVGVGLLWFGWFGFNAGSALAANGTAAAIFLNTLVAGCLG
Mflv_2626|M.gilvum_PYR-GCK TIRPHNLPLTMVGLFLIFTGFFGFLMGCVIYGPGGYATIYGSPTTLSAFA
Mvan_3955|M.vanbaalenii_PYR-1 TIRPHNLPLTMLGLFLIFTGFFGFLMGCVIYGATGYSTIYGSPTTLSAFA
MSMEG_4635|M.smegmatis_MC2_155 TIRPHNLPLTMLGLMLIFTGFFGFLMGCIIYAGDGLTTIYASPTTMSAFA
****:* ::* *:: *::** *. . .. . .
Mb2944c|M.bovis_AF2122/97 MLGWL-----LTERVRDGKATTLGAASGIVAGLVAITPSCSSVNVLGALA
Rv2920c|M.tuberculosis_H37Rv MLGWL-----LTERVRDGKATTLGAASGIVAGLVAITPSCSSVNVLGALA
MAB_0355|M.abscessus_ATCC_1997 MASWL-----LVEKVRHGKPTSFGAASGVVAGLVAITPSCAAVTPIGALI
TH_4321|M.thermoresistible__bu MLGWL-----IVERIRDGRPTSVGAASGIVAGLVAITPACGALSPVGSLF
MMAR_0038|M.marinum_M MLGWL-----SVEQIRDGKPTTFGAASGVVAGLVAITPSCGTVNTLGAAL
MUL_0037|M.ulcerans_Agy99 MLGWL-----AVEQIRDGKPTTFGAASGVVAGLVAITPSCGTVNPLGAAL
MAV_3084|M.avium_104 MLGWL-----SVEQIRDGRPTTFGAASGVVAGLVAITPSCGTVNTLGATV
Mflv_2626|M.gilvum_PYR-GCK FNTLMGLAGGIIGAYLTSRGEPFWTISGGLAGVIAVAAGMDLYHPALTFV
Mvan_3955|M.vanbaalenii_PYR-1 FNTLMGMVGGIIGAYVMSRGEPFWTISGGLAGVIAIAAGMDLYHPALTFV
MSMEG_4635|M.smegmatis_MC2_155 FNTLMGLGGGIIGAYLTSKGEPFWTISGGLCGVIGVAAGMDLYHPTLAFV
: : .: .. : ** :.*::.::.. :
Mb2944c|M.bovis_AF2122/97 VGVSAGVLCALAVGLKFKLGFDDSLDVVGVHLVGGLVGTLLVGLLAAPEA
Rv2920c|M.tuberculosis_H37Rv VGVSAGVLCALAVGLKFKLGFDDSLDVVGVHLVGGLVGTLLVGLLAAPEA
MAB_0355|M.abscessus_ATCC_1997 IGAVTGAVSSLAIELKYRLGYDDSLDVVGVHLVGGVVGTLMIGVVGSAAA
TH_4321|M.thermoresistible__bu LGVIAGALSALAVGLKYRFGYDDSLDVVGVHLVAGLWGTIGIGFFAT---
MMAR_0038|M.marinum_M VGLAAGIVCAFAITVKFKLNYDDSLDVVGVHFIGGVVGVLLIGLLATAVM
MUL_0037|M.ulcerans_Agy99 IGLAAGIVCAFAITVKFKLNYDDSLDVVGVHFIGGVVGVLLIGLLATAVM
MAV_3084|M.avium_104 VGLAAGVVCSFAVGAKLRFNYDDSLDVVGVHFVGGVVGVSLIGLFATAVM
Mflv_2626|M.gilvum_PYR-GCK IALVGGGLIPFIGKLLDKMKIDDVVGAVAVHGGMGFYSLIMAGIFLAGYP
Mvan_3955|M.vanbaalenii_PYR-1 IALVGGGLIPFIGKLLDKMKIDDVVGAVAVHGGMGLYSVIVAGIFLAGYP
MSMEG_4635|M.smegmatis_MC2_155 IAFGAGAVAPFIGRLLEKFKIDDVVGAVSVHGGIGLYSLLMSGIFLHGYP
:. * : .: :: ** :..*.** *. . *..
Mb2944c|M.bovis_AF2122/97 PAINGVAGVSKGLFYGGGFAQLERQALGACSVLVYSGIITLILALILKFT
Rv2920c|M.tuberculosis_H37Rv PAINGVAGVSKGLFYGGGFAQLERQALGACSVLVYSGIITLILALILKFT
MAB_0355|M.abscessus_ATCC_1997 P-----AGVN-GLLYGGGLHQFWLQLVGVVTVLVYSVAVTGLIGLGLRHS
TH_4321|M.thermoresistible__bu ---------ETGLLYGGGIQQLIVQIVIAVVAMAFTAVMTLVIATVLKP-
MMAR_0038|M.marinum_M T------DGARGLFYGGGLGQLGKQALAMVVVALYAFTVSYLLAALIERL
MUL_0037|M.ulcerans_Agy99 T------DGARGLFYGGGLGQLGKQALAMVVVGLYAFTVSYLLAALIERL
MAV_3084|M.avium_104 T------AGPQGLFYGGGVAQLGKQALAIAAVALYAFTVSFLLAKAIDRV
Mflv_2626|M.gilvum_PYR-GCK N------------TDGNPSISFWGQLIGAVTFAALGFLPAYLLSLGLKKA
Mvan_3955|M.vanbaalenii_PYR-1 N------------IGGNPSISFWGQLIGAVVFAALGFIPAYLLSLLLKKV
MSMEG_4635|M.smegmatis_MC2_155 N------------TGGNPSISLWGQAIGAAVFAALGFIPTYVVSLGLKKV
*. .: * : : ::. :
Mb2944c|M.bovis_AF2122/97 IGLRLDAEQESTGIDEAEHAESGYDFAVASGSVLPPRVTVEDSRNGIQER
Rv2920c|M.tuberculosis_H37Rv IGLRLDAEQESTGIDEAEHAESGYDFAVASGSVLPPRVTVEDSRNGIQER
MAB_0355|M.abscessus_ATCC_1997 LGIRSHPQHESDGLDDAEHAESAYELATTRGGGL-----INPGR------
TH_4321|M.thermoresistible__bu LGWRVSEEDEEAGIDAAEHAETAYELV-----------------------
MMAR_0038|M.marinum_M MGFRVSREEEVSGVDLSQHAETAYTEGVYGHQQLHRPFSGGDAIRPRPNI
MUL_0037|M.ulcerans_Agy99 MGFRVSREEEVSGVDFSQHAETAYTEGVYGHQQLHRPFSGGDAIRPRPNI
MAV_3084|M.avium_104 MGFRVSAEDETTGVDLTQHAETAYAEGVHGHLPQRRPGPG-DRLK-----
Mflv_2626|M.gilvum_PYR-GCK GLLRIPPEVEEQGLDLSEVPATPYPEGIPVTEMRASSG-----TAVLVLG
Mvan_3955|M.vanbaalenii_PYR-1 GLLRIPAEVEEQGLDLSEVPATPYPEGIPVTAMKLNG-------SVLVVT
MSMEG_4635|M.smegmatis_MC2_155 GLLRIPKEVEEQGLDLTEVPATPYPEGIPVTAMPVNGNGHAKTNGKLVTT
* : * *:* :: . : *
Mb2944c|M.bovis_AF2122/97 IGQKVEAEPK
Rv2920c|M.tuberculosis_H37Rv IGQKVEAEPK
MAB_0355|M.abscessus_ATCC_1997 ----------
TH_4321|M.thermoresistible__bu ----------
MMAR_0038|M.marinum_M EDA-------
MUL_0037|M.ulcerans_Agy99 EDA-------
MAV_3084|M.avium_104 ----------
Mflv_2626|M.gilvum_PYR-GCK EEVK------
Mvan_3955|M.vanbaalenii_PYR-1 EEVN------
MSMEG_4635|M.smegmatis_MC2_155 AEV-------