For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RQRALVARERTYAHAVAVNNSRRARAARRRKRRVAAAVNDLTDAQWASIKAAWEGCAYCGKTTGTMQRDC VMAISRGGRYTVDNVVPACGACNASKCNDEVTGWLRRKRYDERLFLERYVRIRAELMPAERETALTVAPE AGSLP*
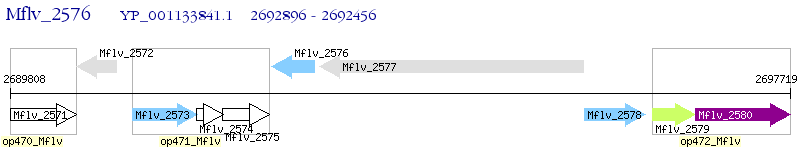
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2576 | - | - | 100% (146) | HNH nuclease |
| M. gilvum PYR-GCK | Mflv_2573 | - | 7e-06 | 45.10% (51) | HNH endonuclease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01254 | - | 2e-05 | 40.00% (55) | hypothetical protein MLBr_01254 |
| M. abscessus ATCC 19977 | MAB_1343 | - | 5e-38 | 65.09% (106) | hypothetical protein MAB_1343 |
| M. marinum M | MMAR_3820 | - | 2e-05 | 41.82% (55) | hypothetical protein MMAR_3820 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4691 | - | 1e-47 | 78.38% (111) | HNH nuclease |
| M. thermoresistible (build 8) | TH_0922 | - | 2e-05 | 42.59% (54) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3745 | - | 2e-05 | 41.82% (55) | hypothetical protein MUL_3745 |
| M. vanbaalenii PYR-1 | Mvan_4058 | - | 1e-58 | 88.43% (121) | HNH nuclease |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2576|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4058|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4691|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1343|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3820|M.marinum_M MAQGKKRRGHRNHGAAASMTGTA--SCLHSVDTHPPNRHETASIWSRRRV
MUL_3745|M.ulcerans_Agy99 MAQGKKRRGHRNHGAAANMTGTA--SCLHSVDTHPPNRHETASIWSRRRV
TH_0922|M.thermoresistible__bu -------------------------------------------------V
MLBr_01254|M.leprae_Br4923 MVQRKNRRSHRSSGAAANLIRAANPSSLHNVDIHPSTCYESGSIWNRRRV
Mflv_2576|M.gilvum_PYR-GCK --------------MRQRA---------LVARERT-YAHAVAVNNSRRAR
Mvan_4058|M.vanbaalenii_PYR-1 --------------MRACVSAGDTPGTWPISRTLTPHALGVAVNNSRRAR
MSMEG_4691|M.smegmatis_MC2_155 ----------------------------------------MAVSRTRSAR
MAB_1343|M.abscessus_ATCC_1997 ------------------------------------------MAGTRRAR
MMAR_3820|M.marinum_M LLLNSTYEPLTALPMRRAIVMVICGKADVVHDDPAGPVIHSATRTIVVPS
MUL_3745|M.ulcerans_Agy99 LLLNSTYEPLTALPMRRAIVMVICGKADVVHDDPAGPVIHSATQTIVVPS
TH_0922|M.thermoresistible__bu LLLNSTYEPLTALTMRRALVMVLCGKADVVHDDPNGPVIHSATRALVMPS
MLBr_01254|M.leprae_Br4923 LLLNSTYEPLTALPTRRAIIMVICGKADVVHVDPAGPVVHSATRSITVPS
.
Mflv_2576|M.gilvum_PYR-GCK AARRRKRRVAAAVNDLTDAQWASIKAAWEGCAYCGKTTGTMQRDCVMAIS
Mvan_4058|M.vanbaalenii_PYR-1 AARRRKRRVAAVVNDLTDEQWAAIRAAWNGCAYCGKTAGTMQRDCVMAIS
MSMEG_4691|M.smegmatis_MC2_155 YARRRKRRLDAVVNDLTPAQWEAIKQAWGGCAYCGATGGPFQKDCVMAIS
MAB_1343|M.abscessus_ATCC_1997 IAKRRRLRVSRADNDLTDAQWSQLVQAWGGCAYCGATGTALQRDCVQPIS
MMAR_3820|M.marinum_M VIQLRTYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTVD--HVVPRS
MUL_3745|M.ulcerans_Agy99 VIQLRTYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTVD--HVVPRS
TH_0922|M.thermoresistible__bu VIRLRTYVRVPYRARVPMTRAALMHRDRFRCAYCGGKADTID--HVVPRS
MLBr_01254|M.leprae_Br4923 VIQLRSYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTVD--HVVPRS
: * :. : : ***** . ..: * . *
Mflv_2576|M.gilvum_PYR-GCK RGGRYTVDNVVPACGACNASKCN---DEVTGWLRRKRYDERLFLERYVRI
Mvan_4058|M.vanbaalenii_PYR-1 RGGRYTVDNVVPACAACNASKCN---DEVTGWLRRKRLDERLFLERYVRI
MSMEG_4691|M.smegmatis_MC2_155 RGGRYTVDNVVPACGSCNASKCN---DEVTGWLRRKRLDERRFLTRYVEI
MAB_1343|M.abscessus_ATCC_1997 RGGRYTQANVVPACGSCNSSKCN---AEVTSWMRRKRLDETAFLRRYVEV
MMAR_3820|M.marinum_M RGGEHSWENCVACCSACNHRKGDKLLTELGWSLRRTPLPPTGQHWRLLST
MUL_3745|M.ulcerans_Agy99 RGGEHSWENCVACCSACNHRKGDKLLTELGWSLRRTPLPPTGQHWRLLST
TH_0922|M.thermoresistible__bu RGGAHSWENCVAACSACNHRKADRLLSELGWTLRAVPRPPKGQHWRLLST
MLBr_01254|M.leprae_Br4923 RGGDHSWENCVACCSTCNHRKGDKLLTELGWVLRRTPVLPTGQHWRLLST
*** :: * *..*.:** * : *: :* * :
Mflv_2576|M.gilvum_PYR-GCK RAELMPAERETALTVAPEAGSLP
Mvan_4058|M.vanbaalenii_PYR-1 RAELV-GEREAGAVDQS------
MSMEG_4691|M.smegmatis_MC2_155 RARLT-----AQFA---------
MAB_1343|M.abscessus_ATCC_1997 TQALA------------------
MMAR_3820|M.marinum_M IKELDPSWARYLG-EGAA-----
MUL_3745|M.ulcerans_Agy99 IKELDPSWARYLG-EGAA-----
TH_0922|M.thermoresistible__bu VKELDPVWVRYLGGEGAA-----
MLBr_01254|M.leprae_Br4923 VKELDPAWARYLG-GGAA-----
*