For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAQRKNGRGHRNHRTAVALPGSSSPTASRALHSVAPAVDAHPVVAESSVWGRRRVLLLNSTYEPLTALPL RRAVIMLMCGKADVVHDDPVGPVIHSATRSIRVPTVIRLRTFVRVPYRARIPMTRAALMHRDRFRCAYCG SKADTVDHVIPRSRGGAHTWENCVAACSACNHRKADKLLSELGWSLHTTPLPPKGQHWRLLSSVKELDPA WVRYLGEGAA
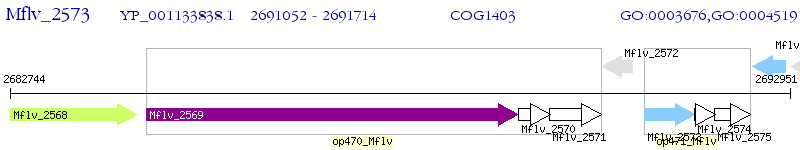
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2573 | - | - | 100% (220) | HNH endonuclease |
| M. gilvum PYR-GCK | Mflv_3897 | - | 2e-07 | 38.33% (60) | HNH endonuclease |
| M. gilvum PYR-GCK | Mflv_2576 | - | 1e-05 | 45.10% (51) | HNH nuclease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2496c | - | 9e-91 | 68.92% (222) | hypothetical protein Mb2496c |
| M. tuberculosis H37Rv | Rv2469c | - | 1e-91 | 69.82% (222) | hypothetical protein Rv2469c |
| M. leprae Br4923 | MLBr_01254 | - | 1e-90 | 70.72% (222) | hypothetical protein MLBr_01254 |
| M. abscessus ATCC 19977 | MAB_1567 | - | 6e-94 | 87.86% (173) | HNH endonuclease precursor |
| M. marinum M | MMAR_3820 | - | 4e-96 | 73.87% (222) | hypothetical protein MMAR_3820 |
| M. avium 104 | MAV_1702 | - | 9e-92 | 71.30% (216) | HNH endonuclease family protein |
| M. smegmatis MC2 155 | MSMEG_4694 | - | 2e-93 | 80.31% (193) | HNH endonuclease family protein |
| M. thermoresistible (build 8) | TH_0922 | - | 2e-84 | 83.83% (167) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3745 | - | 2e-95 | 73.42% (222) | hypothetical protein MUL_3745 |
| M. vanbaalenii PYR-1 | Mvan_4063 | - | 1e-116 | 90.05% (221) | HNH endonuclease |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2496c|M.bovis_AF2122/97 MAHGKKRRGHRSSGVAAGVTGP--ASCLHGVHSHRLASGVETHPPNRHES
Rv2469c|M.tuberculosis_H37Rv MAHGKKRRGHRSSGVAAGVTGP--ASCLHSVHSHRLASGVETHPPNRHES
MMAR_3820|M.marinum_M MAQGKKRRGHRNHGAAASMTGT--ASCLH---------SVDTHPPNRHET
MUL_3745|M.ulcerans_Agy99 MAQGKKRRGHRNHGAAANMTGT--ASCLH---------SVDTHPPNRHET
MAV_1702|M.avium_104 MAQGKKRRRHRSQVAAAGLTGPPTASSLH---------SVETHPPTRPDS
MLBr_01254|M.leprae_Br4923 MVQRKNRRSHRSSGAAANLIRAANPSSLH---------NVDIHPSTCYES
TH_0922|M.thermoresistible__bu --------------------------------------------------
Mflv_2573|M.gilvum_PYR-GCK MAQRKNGRGHRNHRTAVALPGSSSPTASRALHS--VAPAVD-AHPV-VAE
Mvan_4063|M.vanbaalenii_PYR-1 MAQRKNGRGHRHHRTAAGTSGSTSATTSRALHS--VAPSVDTLTPA-PQD
MAB_1567|M.abscessus_ATCC_1997 MPPQQSN-----------------------------------------DA
MSMEG_4694|M.smegmatis_MC2_155 MPNRPLHS-------------------------------VEPLASA-EPD
Mb2496c|M.bovis_AF2122/97 ASIWNRRRVLLLNSTYEPLTALSMRRAIVMVICGKADVVHEDPSGPVIHS
Rv2469c|M.tuberculosis_H37Rv ASIWNRRRVLLLNSTYEPLTALSMRRAIVMVICGKADVVHEDPSGPVIHS
MMAR_3820|M.marinum_M ASIWSRRRVLLLNSTYEPLTALPMRRAIVMVICGKADVVHDDPAGPVIHS
MUL_3745|M.ulcerans_Agy99 ASIWSRRRVLLLNSTYEPLTALPMRRAIVMVICGKADVVHDDPAGPVIHS
MAV_1702|M.avium_104 TSIWSRRRVLLLNSTYEPLTALPMRRAIVMVICGKADVVHHDPAGPVIHS
MLBr_01254|M.leprae_Br4923 GSIWNRRRVLLLNSTYEPLTALPTRRAIIMVICGKADVVHVDPAGPVVHS
TH_0922|M.thermoresistible__bu --------VLLLNSTYEPLTALTMRRALVMVLCGKADVVHDDPNGPVIHS
Mflv_2573|M.gilvum_PYR-GCK SSVWGRRRVLLLNSTYEPLTALPLRRAVIMLMCGKADVVHDDPVGPVIHS
Mvan_4063|M.vanbaalenii_PYR-1 GSVWGRRRVLLLNSTYEPLTALPLRRAVIMLMCGKADVVHDDPVGPVIHS
MAB_1567|M.abscessus_ATCC_1997 GSLWGRRRVLLLNSTYEPLTALPMRRAVIMLLCGKADVVHDDPAAPIIHS
MSMEG_4694|M.smegmatis_MC2_155 ASLWGRRRVLLLNATYEPLTALPLRRAVIMVVCGKADVVHDDPTSPVIHS
*****:********. ***::*::******** ** .*::**
Mb2496c|M.bovis_AF2122/97 STRSILVPSVIQLRSYVRVPYRARVPMTRAALMHRDRFCCAYCGGKADTV
Rv2469c|M.tuberculosis_H37Rv ATRSILVPSVIQLRSYVRVPYRARVPMTRAALMHRDRFCCAYCGGKADTV
MMAR_3820|M.marinum_M ATRTIVVPSVIQLRTYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTV
MUL_3745|M.ulcerans_Agy99 ATQTIVVPSVIQLRTYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTV
MAV_1702|M.avium_104 ATSSIVMPSVIQLRSYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTV
MLBr_01254|M.leprae_Br4923 ATRSITVPSVIQLRSYVRVPYRARVPMTRAALMHRDRFCCAYCGAKADTV
TH_0922|M.thermoresistible__bu ATRALVMPSVIRLRTYVRVPYRARVPMTRAALMHRDRFRCAYCGGKADTI
Mflv_2573|M.gilvum_PYR-GCK ATRSIRVPTVIRLRTFVRVPYRARIPMTRAALMHRDRFRCAYCGSKADTV
Mvan_4063|M.vanbaalenii_PYR-1 ATRSIAVPTVIRLRTFVRVPYRARIPMTRAALMHRDRFRCAYCSAKADTV
MAB_1567|M.abscessus_ATCC_1997 ATTSVAVPSVIRLRTFVRVPYRARVPMTRAALMHRDRFRCAYCGGRADTI
MSMEG_4694|M.smegmatis_MC2_155 ASRSIVVPSVIRLRTYVRVPYRARVPMTRAALMHRDRFRCAYCGGRADTV
:: :: :*:**:**::********:************* ****..:***:
Mb2496c|M.bovis_AF2122/97 DHVVPRSRGGAHSWENCVACCSPCNHRKGDRLLTELGWALRRAPLPPTGP
Rv2469c|M.tuberculosis_H37Rv DHVVPRSRGGAHSWENCVACCSPCNHRKGDRLLTELGWALRRAPLPPTGP
MMAR_3820|M.marinum_M DHVVPRSRGGEHSWENCVACCSACNHRKGDKLLTELGWSLRRTPLPPTGQ
MUL_3745|M.ulcerans_Agy99 DHVVPRSRGGEHSWENCVACCSACNHRKGDKLLTELGWSLRRTPLPPTGQ
MAV_1702|M.avium_104 DHVVPRSRGGDHSWENCVACCSTCNHRKGDKLLNELGWTLRRPPLPPTGQ
MLBr_01254|M.leprae_Br4923 DHVVPRSRGGDHSWENCVACCSTCNHRKGDKLLTELGWVLRRTPVLPTGQ
TH_0922|M.thermoresistible__bu DHVVPRSRGGAHSWENCVAACSACNHRKADRLLSELGWTLRAVPRPPKGQ
Mflv_2573|M.gilvum_PYR-GCK DHVIPRSRGGAHTWENCVAACSACNHRKADKLLSELGWSLHTTPLPPKGQ
Mvan_4063|M.vanbaalenii_PYR-1 DHVIPRSRGGAHSWENCVAACSACNHRKADRLLSELGWSLHTTPLPPKGQ
MAB_1567|M.abscessus_ATCC_1997 DHVIPRSKGGAHSWENCVACCSSCNHRKADRLLAELGWSLHTTPMPPKGQ
MSMEG_4694|M.smegmatis_MC2_155 DHVVPRSRGGEHSWENCVAACATCNHRKADRLLTELGWTLRCVPMPPKGQ
***:***:** *:******.*:.*****.*:** **** *: * *.*
Mb2496c|M.bovis_AF2122/97 HWRLLSAVKELDPSWARYLG-EGAA
Rv2469c|M.tuberculosis_H37Rv HWRLLSAVKELDPSWARYLG-EGAA
MMAR_3820|M.marinum_M HWRLLSTIKELDPSWARYLG-EGAA
MUL_3745|M.ulcerans_Agy99 HWRLLSTIKELDPSWARYLG-EGAA
MAV_1702|M.avium_104 HWRLLSTVKELDPAWARYLG-EGAA
MLBr_01254|M.leprae_Br4923 HWRLLSTVKELDPAWARYLG-GGAA
TH_0922|M.thermoresistible__bu HWRLLSTVKELDPVWVRYLGGEGAA
Mflv_2573|M.gilvum_PYR-GCK HWRLLSSVKELDPAWVRYLG-EGAA
Mvan_4063|M.vanbaalenii_PYR-1 HWRLLSSVKELDPAWVRYLG-EGAA
MAB_1567|M.abscessus_ATCC_1997 HWRLLSSVKELDPAWVRYLG-EGAA
MSMEG_4694|M.smegmatis_MC2_155 HWRLLSTVKELDPAWVRYLG-EGAA
******::***** *.**** ***