For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDAHDVAGIELTDNILWLLKQAFYFSLTSVNEAVSEHGVSTAQIGVLRQLANEPGLSGAELARRLLISPQ GVQLALTALERRGLVERKQNPSHGRILQAFLTDQGRDVAAAVVQDAIAAHDKVFGVLSKAEQKTLRELLG RVVEQGTGHELFADHVDT
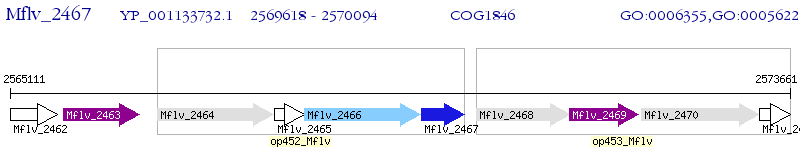
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2467 | - | - | 100% (158) | MarR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1190 | - | 1e-06 | 29.31% (116) | MarR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1731 | - | 8e-06 | 28.07% (114) | MarR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2911 | - | 2e-12 | 35.40% (113) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv2887 | - | 2e-12 | 35.40% (113) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02140 | - | 9e-05 | 28.77% (73) | putative MarR-family protein |
| M. abscessus ATCC 19977 | MAB_2472c | - | 2e-12 | 29.92% (127) | MarR family transcriptional regulator |
| M. marinum M | MMAR_1822 | - | 9e-12 | 34.15% (123) | transcriptional regulatory protein |
| M. avium 104 | MAV_3964 | - | 1e-66 | 83.22% (149) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | MSMEG_0296 | - | 8e-64 | 79.19% (149) | transcriptional regulator, MarR family protein |
| M. thermoresistible (build 8) | TH_0263 | - | 1e-57 | 71.05% (152) | transcriptional regulator, MarR family protein |
| M. ulcerans Agy99 | MUL_2070 | - | 5e-11 | 33.33% (123) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4187 | - | 9e-78 | 91.08% (157) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2911|M.bovis_AF2122/97 ------MGLADDAPLGYLLYRVGAVLRPEVSAALSPLGLTLPEFVCLRML
Rv2887|M.tuberculosis_H37Rv ------MGLADDAPLGYLLYRVGAVLRPEVSAALSPLGLTLPEFVCLRML
MMAR_1822|M.marinum_M ------MGQAEEAPLGFLLYRAAAVLRPEVSTALSPLGLTLPEFVCLRIL
MUL_2070|M.ulcerans_Agy99 ---MVDMGQAEEAPLGFLLYRAAAVLRPEVSTALSPLGLTLPEFVCLRIL
MAB_2472c|M.abscessus_ATCC_199 --------------MGILVSQMYRLLWSRASVIVRDFGVTVPQMECLAVL
Mflv_2467|M.gilvum_PYR-GCK MDAHDVAGIELTDNILWLLKQAFYFSLTSVNEAVSEHGVSTAQIGVLRQL
Mvan_4187|M.vanbaalenii_PYR-1 MDARDVTGIELTDNILWLLKQAFYFSLTTVNEAISEHGVSTAQIGVLRQL
MAV_3964|M.avium_104 --------MELTDNILWLLKQAFYYSLTTINEAMREHGVSTAQIGVLRQL
MSMEG_0296|M.smegmatis_MC2_155 --------MELTDNTLWWLKQAFYFSLTAVNDAVKDHGVSTAQIGVLRQL
TH_0263|M.thermoresistible__bu --VADDLAMEFGDNLLWHLKQAWYFSLTAVNDAVSKHGVSTAQIGVLRQL
MLBr_02140|M.leprae_Br4923 -------MLDSDARLASDLSLAVMRLARQLRFRNPQSPVSLSQLSALTTL
: :: .:: * *
Mb2911|M.bovis_AF2122/97 SQSPGLSSAELARHASVTPQAMNTVLRKLEDAGAVARPASVSSGRSLPAT
Rv2887|M.tuberculosis_H37Rv SQSPGLSSAELARHASVTPQAMNTVLRKLEDAGAVARPASVSSGRSLPAT
MMAR_1822|M.marinum_M SASPGLSSAELARHTNVTPQAMNTVLRKLEEVGAVSRPASVPSGRALPAT
MUL_2070|M.ulcerans_Agy99 SASPGLSSAELARHTNVTSQAMNTVLRKLEEVGAVSRPASVPSGRALPAT
MAB_2472c|M.abscessus_ATCC_199 SAQPGISNVEIAAELRITPQAVSLVQRSLEEAGLVHRSRRQADARVLTTE
Mflv_2467|M.gilvum_PYR-GCK ANEPGLSGAELARRLLISPQGVQLALTALERRGLVERKQNPSHGRILQAF
Mvan_4187|M.vanbaalenii_PYR-1 ANEPGLSGAELARRLLISPQGVQLALTALERQGLVERKQNPAHGRILQAF
MAV_3964|M.avium_104 ANEPGLSGAELARRLLISPQGVQLALTALERRGLVERKRDPQHARILQAH
MSMEG_0296|M.smegmatis_MC2_155 SNEPGLSGAELARRLLITPQGVQLALTALEKRGLVERKQDPQHARILQVF
TH_0263|M.thermoresistible__bu SNEPGLSGAELARRLLITPQGVQLALKSLEQRGLIERRPDPQHARILKVY
MLBr_02140|M.leprae_Br4923 ANEGAMTPGALAIRERVRPPSMTRVIASLADMGFVDREPHPVDGRQVLVS
: . .:: :* . : . .: . * * : * .* : .
Mb2911|M.bovis_AF2122/97 LTARGRALAKRAEAVVRAADARVLARLTAPQQREFKRMLEKLGSD-----
Rv2887|M.tuberculosis_H37Rv LTARGRALAKRAEAVVRAADARVLARLTAPQQREFKRMLEKLGSD-----
MMAR_1822|M.marinum_M LTAPGRALLKRAEVVVRGADDRILAKLTPAEQREFKRMLEMLGTE-----
MUL_2070|M.ulcerans_Agy99 LTAPGRALLKRAEVVVRGADDRILAKLTPAEQREFKRMLEMLGTE-----
MAB_2472c|M.abscessus_ATCC_199 LTAKGRKLFERADATLREDDEEILANMTERDRVQLRRLLGGVIETISAR-
Mflv_2467|M.gilvum_PYR-GCK LTDQGRDVAAAVVQDAIAAHDKVFGVLSKAEQKTLRELLGRVVEQGTGHE
Mvan_4187|M.vanbaalenii_PYR-1 LTDKGRKVAAAVLHDAIAAHDRVFGVLSKAEQQTLRELLGRVVEQGTGHE
MAV_3964|M.avium_104 LTGEGRKVAETVVNDAIAAHQEVFGVLTEQEQQTLRELLGRVVEKGTGHE
MSMEG_0296|M.smegmatis_MC2_155 LTEEGRAVASSVVSDAIAAHDRVFGVLSRDEQEQLRDLLRRVIEQGTGHT
TH_0263|M.thermoresistible__bu LTREGRSVAKAVVGDAIAAHEAVFGVLTPEEQRTLRDLLARVVEKGTGHT
MLBr_02140|M.leprae_Br4923 VSQSGAELVKAARRARQEWLVERLMTLNSDKRDILRNAADLILELIDESP
:: * : . : :. .: :: :
Mb2911|M.bovis_AF2122/97 ---------
Rv2887|M.tuberculosis_H37Rv ---------
MMAR_1822|M.marinum_M ---------
MUL_2070|M.ulcerans_Agy99 ---------
MAB_2472c|M.abscessus_ATCC_199 ---------
Mflv_2467|M.gilvum_PYR-GCK LFADHVDT-
Mvan_4187|M.vanbaalenii_PYR-1 LYTDHVDP-
MAV_3964|M.avium_104 LFTDHVDG-
MSMEG_0296|M.smegmatis_MC2_155 VQADHVDPM
TH_0263|M.thermoresistible__bu LADDHITD-
MLBr_02140|M.leprae_Br4923 ---------