For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTQTVTDDVAAEATRDTGPVVLDIVIPVHNEQAALADSVRRVHRYLNESVPLRARITIADNASVDETPRV AAQLAADLPGVRVVRLEEKGRGRALREVWTRSDAAVLVYMDVDLSTDLAALAPLVAPLISGHSDLAIGTR LARSARVQRGPKREIISRCYNMILKSTLSAGFSDAQCGFKAIRADVAAQLLPYVEDTGWFFDTELLILAE RSGLRIHEVPVDWVDDPDSRVDIVSTAAADLKGIGRLLRGFAGGAIPVNTIAAQLGKSGGPRRSLLRQAV RFGAVGVVSTVAYTVLFLLMQGWAGAQPANVIALLLTALGNTAANRRFTFGVGGRHRRGRHHVEGLIVFA IALALTSGALALLHAVAESPHHLVGLAVLVVANLLATVVRFVLLRGWVFHPRRTTTGGTTR
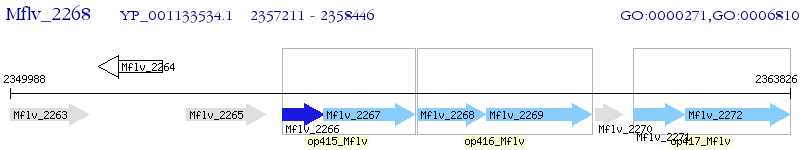
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2268 | - | - | 100% (411) | glycosyl transferase family protein |
| M. gilvum PYR-GCK | Mflv_3118 | - | 4e-12 | 29.19% (209) | dolichyl-phosphate beta-D-mannosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2077c | ppm1 | 1e-12 | 28.71% (209) | polyprenol-monophosphomannose synthase Ppm1 |
| M. tuberculosis H37Rv | Rv2051c | ppm1 | 1e-12 | 28.71% (209) | polyprenol-monophosphomannose synthase Ppm1 |
| M. leprae Br4923 | MLBr_00207 | - | 2e-07 | 26.23% (244) | putative glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_0926 | - | 1e-122 | 59.25% (400) | glycosyltransferase |
| M. marinum M | MMAR_5093 | ppm1_2 | 2e-14 | 29.67% (209) | polyprenol-monophosphomannose synthase Ppm1_2 |
| M. avium 104 | MAV_0525 | - | 6e-06 | 22.78% (237) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_4987 | - | 1e-166 | 75.82% (397) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_0733 | - | 6e-10 | 30.51% (177) | Polyprenol-monophosphomannose synthase Ppm1 |
| M. ulcerans Agy99 | MUL_4169 | ppm1_2 | 1e-13 | 29.19% (209) | polyprenol-monophosphomannose synthase Ppm1_2 |
| M. vanbaalenii PYR-1 | Mvan_4426 | - | 0.0 | 83.66% (404) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 MKLGAWVAAQLPTTRTAVRTRLTRLVVSIVAGLLLYASFPPRNCWWAAVV
Rv2051c|M.tuberculosis_H37Rv MKLGAWVAAQLPTTRTAVRTRLTRLVVSIVAGLLLYASFPPRNCWWAAVV
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 ALALLAWVLTHRATTPVGGLGYGLLFGLVFYVSLLPWIGELVGPGPWLAL
Rv2051c|M.tuberculosis_H37Rv ALALLAWVLTHRATTPVGGLGYGLLFGLVFYVSLLPWIGELVGPGPWLAL
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 ATTCALFPGIFGLFAVVVRLLPGWPIWFAVGWAAQEWLKSILPFGGFPWG
Rv2051c|M.tuberculosis_H37Rv ATTCALFPGIFGLFAVVVRLLPGWPIWFAVGWAAQEWLKSILPFGGFPWG
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 SVAFGQAEGPLLPLVQLGGVALLSTGVALVGCGLTAIALEIEKWWRTGGQ
Rv2051c|M.tuberculosis_H37Rv SVAFGQAEGPLLPLVQLGGVALLSTGVALVGCGLTAIALEIEKWWRTGGQ
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 GDAPPAVVLPAACICLVLFAAIVVWPQVRHAGSGSGGEPTVTVAVVQGNV
Rv2051c|M.tuberculosis_H37Rv GDAPPAVVLPAACICLVLFAAIVVWPQVRHAGSGSGGEPTVTVAVVQGNV
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PRLGLDFNAQRRAVLDNHVEETLRLAADVHAGLAQQPQFVIWPENSSDID
Rv2051c|M.tuberculosis_H37Rv PRLGLDFNAQRRAVLDNHVEETLRLAADVHAGLAQQPQFVIWPENSSDID
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PFVNPDAGQRISAAAEAIGAPILIGTLMDVPGRPRENPEWTNTAIVWNPG
Rv2051c|M.tuberculosis_H37Rv PFVNPDAGQRISAAAEAIGAPILIGTLMDVPGRPRENPEWTNTAIVWNPG
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 TGPADRHDKAIVQPFGEYLPMPWLFRHLSGYADRAGHFVPGNGTGVVRIA
Rv2051c|M.tuberculosis_H37Rv TGPADRHDKAIVQPFGEYLPMPWLFRHLSGYADRAGHFVPGNGTGVVRIA
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 GVPVGVATCWEVIFDRAPRKSILGGAQLLTVPSNNATFNKTMSEQQLAFA
Rv2051c|M.tuberculosis_H37Rv GVPVGVATCWEVIFDRAPRKSILGGAQLLTVPSNNATFNKTMSEQQLAFA
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 KVRAVEHDRYVVVAGTTGISAVIAPDGGELIRTDFFQPAYLDSQVRLKTR
Rv2051c|M.tuberculosis_H37Rv KVRAVEHDRYVVVAGTTGISAVIAPDGGELIRTDFFQPAYLDSQVRLKTR
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4426|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4987|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0926|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 LTPATRWGPILQWILVGAAAAVVLVAMRQNGWFPRPRRSEPKGENDDSDA
Rv2051c|M.tuberculosis_H37Rv LTPATRWGPILQWILVGAAAAVVLVAMRQNGWFPRPRRSEPKGENDDSDA
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK --------------------------MTQTVTDD--------------VA
Mvan_4426|M.vanbaalenii_PYR-1 ---------------MSTRPENTMDAMTDLALQSGLRCAERP-FGARPNA
MSMEG_4987|M.smegmatis_MC2_155 --------------------------MTELAVDPETPVEQRFRFASRPNA
MAB_0926|M.abscessus_ATCC_1997 --------------------------MTSVMQPIDT-----------PTA
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PPGRSEASGPPALSESDDELIQPEQGGRHSSGFGRHRATSRSYMTTGQPA
Rv2051c|M.tuberculosis_H37Rv PPGRSEASGPPALSESDDELIQPEQGGRHSSGFGRHRATSRSYMTTGQPA
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M -------------------------------------------MTHERDP
MUL_4169|M.ulcerans_Agy99 -------------------------------------------MTHERDP
Mflv_2268|M.gilvum_PYR-GCK AEATRDTGPVVLDIVIPVHNEQAALADSVRRVHRYLNESVPLRARITIAD
Mvan_4426|M.vanbaalenii_PYR-1 ALVARATGAPVLDVVVPVYNEQAALADSVHRLHRYLSESVPFPARITIAD
MSMEG_4987|M.smegmatis_MC2_155 AQVARAAGVPVLDVVVPVFNEQAALAHSVRRLHGYLQENFALPTRITIAD
MAB_0926|M.abscessus_ATCC_1997 PTAIRAT---TLDIVIPVYNEEHDLEPCVRRLHAFLADEVPYSARITIAD
MLBr_00207|M.leprae_Br4923 --MGIETCYPGVWIVIPAFNEAAIIGEVVTDVRAVFDH-------VVCVD
MAV_0525|M.avium_104 --------------MIPAFNEAAVIGEVIADVRSVFDN-------VVCVD
Mb2077c|M.bovis_AF2122/97 PPAPGNRPSQRVLVIIPTFNERENLPVIHRRLTQACPA-----VHVLVVD
Rv2051c|M.tuberculosis_H37Rv PPAPGNRPSQRVLVIIPTFNERENLPVIHRRLTQACPA-----VHVLVVD
TH_0733|M.thermoresistible__bu -------------------------------------------VHVLIVD
MMAR_5093|M.marinum_M HEHSGTGRSRRVLVIIPTYNERENLPLIHRRVKDACPD-----VHVLVVD
MUL_4169|M.ulcerans_Agy99 HENSGTGRSRRVLVIIPTYNERENLPLIHRRVKDACPD-----VHVLVVD
: .*
Mflv_2268|M.gilvum_PYR-GCK NASVDETPRVAAQLAADLPGVRVVRLEEKGRGRALREVW-----TRSDAA
Mvan_4426|M.vanbaalenii_PYR-1 NASVDDTPRIAAELAAELSGVRVVRLEEKGRGRALHQVW-----AESDAA
MSMEG_4987|M.smegmatis_MC2_155 NASTDATPQIAARLAEELPGVRVVRLEEKGRGRALHAVW-----STSDAP
MAB_0926|M.abscessus_ATCC_1997 NASTDSTLKIAHRLSRELVGVDVLHLEEKGRGRALAAAW-----LSSDAE
MLBr_00207|M.leprae_Br4923 DGSADGTGEIALRAGAHLVRHPVNLGQGAAIQTGVEYAR-----RQPAAQ
MAV_0525|M.avium_104 DGSTDDTGEIARRAGAHLVRHPINLGQGAAIQTGVEYAR-----RQPGAQ
Mb2077c|M.bovis_AF2122/97 DSSPDGTGQLADELAQADPGRTHVMHRTAKNGLGAAYLAGFAWGLSREYS
Rv2051c|M.tuberculosis_H37Rv DSSPDGTGQLADELAQADPGRTHVMHRTAKNGLGAAYLAGFAWGLSREYS
TH_0733|M.thermoresistible__bu DGSPDGTGELADELALADPDRVHVMHRASKAGLGAAYLAGFRWGLNRQYS
MMAR_5093|M.marinum_M DESPDGTGQLADELAAADPGCTYVLHRNVKDGLAAAYLQGFVWGMQRNYS
MUL_4169|M.ulcerans_Agy99 DESPDGTGQLADELAAADLGCTYVLHRNVKDGLAAAYLQGFVWGMQRNYS
: * * * .:* . . . .
Mflv_2268|M.gilvum_PYR-GCK VLVYMDVDLSTDLAALAPLVAPLISGHSDLAIGTRLARSARVQ------R
Mvan_4426|M.vanbaalenii_PYR-1 VLVYMDVDLSTDLAALAPLVAPLISGHSDLAIGTRLARGARVR------R
MSMEG_4987|M.smegmatis_MC2_155 VLAYMDVDLSTDLAALAPLVAPLISGHSDLAIGTRLGRGSRVI------R
MAB_0926|M.abscessus_ATCC_1997 VVAYCDVDLSTDLNALMPLVAPLISGHSDVAYGSRLNRNSRVV------R
MLBr_00207|M.leprae_Br4923 VFATFDADGQHRVKDLAALVDRLGAHDVDVVIGTRFGRLDGGRLPILKGP
MAV_0525|M.avium_104 VFATFDADGQHRVKDLAAMVNRLRVGDVDVVIGTRFGTRQGAR------P
Mb2077c|M.bovis_AF2122/97 VLVEMDADGSHAPEQLQRLLDAVDAG-ADLAIGSRYVAGGTVRN-----W
Rv2051c|M.tuberculosis_H37Rv VLVEMDADGSHAPEQLQRLLDAVDAG-ADLAIGSRYVAGGTVRN-----W
TH_0733|M.thermoresistible__bu VLVEMDADGSHAPEELHRLLDAVDAG-ADLAIGSRYVDGGQVRN-----W
MMAR_5093|M.marinum_M VLVEMDADGSHAPEQLHRLLEAVEAG-ADLAIGSRYVRGGTVRN-----W
MUL_4169|M.ulcerans_Agy99 VLVEMDADGSHAPEQLHRLLEAVEAG-ADLAIGSRYVRGGTVRN-----W
*.. *.* . * :: : *:. *:*
Mflv_2268|M.gilvum_PYR-GCK GPKREIISRCYNMILKSTLSAGFSDAQCGFKAIRADVAAQLLPY-VEDTG
Mvan_4426|M.vanbaalenii_PYR-1 GPKREIISRCYNLILKSTLSAGFSDAQCGFKAIRADVAAQLLPY-VEDTG
MSMEG_4987|M.smegmatis_MC2_155 GAKREFISRCYNLILKSTLAAKFSDAQCGFKAIRADVAKSLLPH-VVDTG
MAB_0926|M.abscessus_ATCC_1997 GPKREFISRTYNLILHASLQVRFSDAQCGFKAIRTDVARQLLPL-VEDKE
MLBr_00207|M.leprae_Br4923 PFLKRVVLRTAARLSRRGRRLGLTDTNNGLRVFNKKVADGLD---ITMTG
MAV_0525|M.avium_104 PLVKRMVLQTAARLSRRGRRLGLTDTNNGLRVFNKKVADGLD---ITMSG
Mb2077c|M.bovis_AF2122/97 PWRRLVLSKTANTYSRLALGIGIHDITAGYRAYRREALEAIDLDGVDSKG
Rv2051c|M.tuberculosis_H37Rv PWRRLVLSKTANTYSRLALGIGIHDITAGYRAYRREALEAIDLDGVDSKG
TH_0733|M.thermoresistible__bu PRRRLLLSRTANTYSRLLLGVDIRDITAGYRAYRREVLEKIDLDAVDSKG
MMAR_5093|M.marinum_M PRRRRVLSKAANTYSRLILGVGVYDITGGYRAYRREVLEALDLDTLESRG
MUL_4169|M.ulcerans_Agy99 PRRRRVLSKAANTYSRLILGVGVYDTTGGYRAYRREVLEALDLDTLESRG
: .: : : . * * :. . .. : :
Mflv_2268|M.gilvum_PYR-GCK WFFDTELLILAERSGLRIHEVPVDWVDDPDSRVDIVSTAAADLKGIGRLL
Mvan_4426|M.vanbaalenii_PYR-1 WFFDTELLVLAERSGLRIHEVPVDWVDDPDSRVDIVATAAADLKGIGRLL
MSMEG_4987|M.smegmatis_MC2_155 WFFDTELLVLAERSGLRIHEVPVDWVDDPDSRVDIVATAAADLKGIGRLL
MAB_0926|M.abscessus_ATCC_1997 WFFDTELLVLAERVGLRIHEVPVDWVDDPDSRVDIVDTVRKDLLGIWRLG
MLBr_00207|M.leprae_Br4923 MSHANEFVMLIADNHWRVDEVPVEVLYTEYSKS----KGQPLLNGVNIIF
MAV_0525|M.avium_104 MSHANEFVMLIDENHWRVVEEPIEVLYTEYSRS----KGQPLLNGVNIIF
Mb2077c|M.bovis_AF2122/97 YCFQIDLTWRTVSNGFVVTEVPITFTERELGVSK--MSGSNIREALVKVA
Rv2051c|M.tuberculosis_H37Rv YCFQIDLTWRTVSNGFVVTEVPITFTERELGVSK--MSGSNIREALVKVA
TH_0733|M.thermoresistible__bu YCFQIDLTWRAINHGFKVVEVPITFTERELGQSK--MSGSNIREAIFKVA
MMAR_5093|M.marinum_M YCFQVEMTWRSLNAGFAVVEVPITFTEREVGASK--MSGSNIGEGIVQVA
MUL_4169|M.ulcerans_Agy99 YCFQVEMTWRSLNAGFAVVEVPITFTEGEVGASK--MSGSNIGEGIAQVA
. :: : * *: . . .: :
Mflv_2268|M.gilvum_PYR-GCK RGFAGGAIPVNTIAAQLGKS---GGPRRSLLRQAVRFGAVGVVSTVAYTV
Mvan_4426|M.vanbaalenii_PYR-1 RGFASGAIPVNAIAAQLGNSR-AAAPPRSLLRQAVRFGTVGIASTLAYIL
MSMEG_4987|M.smegmatis_MC2_155 RGFANGSIPVNTIAAQLGSTR-RTAPPGSLLRQVVRFGAIGVVSTLAYLL
MAB_0926|M.abscessus_ATCC_1997 RALMLGTLPLEELRHSLGREPLVDGVPAGMVGQLVRFGIIGVASTLAYAV
MLBr_00207|M.leprae_Br4923 DGFLRGRMPK----------------------------------------
MAV_0525|M.avium_104 DGFLRGRIPR----------------------------------------
Mb2077c|M.bovis_AF2122/97 RWGIEGRLSRSDHARARPDIARPGAGGSRVSRADVTE-------------
Rv2051c|M.tuberculosis_H37Rv RWGIEGRLSRSDHARARPDIARPGAGGSRVSRADVTE-------------
TH_0733|M.thermoresistible__bu EWGIRGRLDR---ARG---IVR----------------------------
MMAR_5093|M.marinum_M RWGVKDRLRGLRTARISLAAQRFAAAVWTAIRERPARRPAWRPAASIRSH
MUL_4169|M.ulcerans_Agy99 RWGVKDRLRGLRTARISLAAQRFAAAVWTAIRERRARRPAWRPAASIRSH
. :
Mflv_2268|M.gilvum_PYR-GCK LFLLMQGWAGAQPANVIALLLTALGNTAANRRFTFGVGGRHRRGRHHVEG
Mvan_4426|M.vanbaalenii_PYR-1 LFMLLQGWAGAQLANLIALLLTAIGNTAANRRFTFGIGGRPHLARNHVEG
MSMEG_4987|M.smegmatis_MC2_155 LFSLMRAGLGAQAANLIALLLTAIGNTAANRRFTFGIAGTGNVARHHLEG
MAB_0926|M.abscessus_ATCC_1997 LFLALHGLAGDQAANFLALLITAVLNTSANRFFTFGVRGRRDAAKHQFQG
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 --------------------------------------------------
Rv2051c|M.tuberculosis_H37Rv --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M SVHGHAASSAGSSPPTVARSCPPHTS------------------------
MUL_4169|M.ulcerans_Agy99 SVHGHAASSAGSSPPTVARSCPPHTS------------------------
Mflv_2268|M.gilvum_PYR-GCK LIVFAIALALTSGALALLHAVAESPHHLVGLAVLVVANLLATVVRFVLLR
Mvan_4426|M.vanbaalenii_PYR-1 LIVFGIALAITSGALGLLHAATASPHHLVELGVLVTANLLATVVRFVLLR
MSMEG_4987|M.smegmatis_MC2_155 LTVFGIALTITSGSLYLLHVMTPQPHRAVELGVLVAANLIATVVRFVLLR
MAB_0926|M.abscessus_ATCC_1997 LVIFGIGLALTSGSLVAMHHLVPDASKHVLLIVLTVANLVATLIRFVGLR
MLBr_00207|M.leprae_Br4923 --------------------------------------------------
MAV_0525|M.avium_104 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 --------------------------------------------------
Rv2051c|M.tuberculosis_H37Rv --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
MMAR_5093|M.marinum_M --------------------------------------------------
MUL_4169|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2268|M.gilvum_PYR-GCK GWVFHPRRTTTGGTTR-----
Mvan_4426|M.vanbaalenii_PYR-1 GWVFHPRRTHTEGSTQ-----
MSMEG_4987|M.smegmatis_MC2_155 GWVFHPARTRRPSQAPSQIED
MAB_0926|M.abscessus_ATCC_1997 -WVFRSANSR-----------
MLBr_00207|M.leprae_Br4923 ---------------------
MAV_0525|M.avium_104 ---------------------
Mb2077c|M.bovis_AF2122/97 ---------------------
Rv2051c|M.tuberculosis_H37Rv ---------------------
TH_0733|M.thermoresistible__bu ---------------------
MMAR_5093|M.marinum_M ---------------------
MUL_4169|M.ulcerans_Agy99 ---------------------