For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALDRYIGRVGALAVALGIGSAVAAMPGQASAKPDADGGSGMSETSVSGPARAGSGTDATSRSGKPAQSER SAATKPSGGSDPPDRSPREVAVTEETTSGETADDEAEPVEETRDTVEIVPPTEPVTPASAVETEDEPDSP ASSSLLWFLGAAGRRQTDQQAGPDRDAASQTAAQAEAPIAATPVPGDASASPIIGVDGTVYQVTRTSSAG GQFSTVTILDNTGQIVSSGEVRGTVMDAVARPDGTLVVFSRRFISTTVSTVSSDGTVTPIANLLGTPLNP PRVGPDGALYFGAGLPDLLSPVGANVGYRTYRISPRNTVRAFAFDTDVAIADDGTAYLVSRRYGISVLRV IPTAGLGRTTLLPLGTDPSAPVLGPDGTAHVTAGVTWFGSPQTRVYTATGDSIAAHTVAGLPAGAVVGAD GVYLATFVEPGSEGPGTSYVSRLTAGGVGTSGLIEGRIAEFQVTAEGTVFAPIDGAGAETPVAVIDRDGT VHTVLLPGTLVVGRRAIRDGGTYNVEDVGYVNYATGGREYVAVLSADGTVARTIELPEGATGGSVFYSPD GAAFEMLEYRDAQGQTTFRQILALASTTYTDKVPGGRFTYVRDTVVFGPDGVGYLLTGSVSTQIGASQDL DVLGFNAAGETVSRAGGLTNPVTTYENNPYRTVLAFGRDGTAYVTLYFTPDDPGVYAVTATGARKVVDLE YVQYRQGFPPTFGLDGTGYVTSAELLSGDADGIALVTVFSPVTDL*
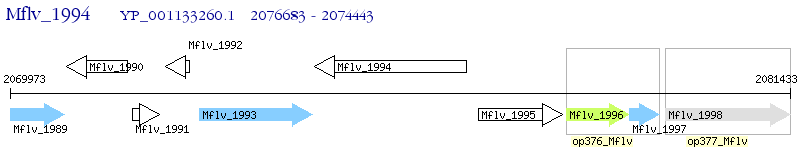
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1994 | - | - | 100% (746) | hypothetical protein Mflv_1994 |
| M. gilvum PYR-GCK | Mflv_5229 | - | 3e-14 | 32.68% (205) | peptidase S15 |
| M. gilvum PYR-GCK | Mflv_3001 | - | 3e-14 | 26.01% (446) | chitinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1842c | - | 4e-05 | 28.35% (194) | dehydrogenase |
| M. tuberculosis H37Rv | Rv1812c | - | 4e-05 | 28.35% (194) | dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4293 | - | 3e-05 | 23.66% (186) | putative Fe-S oxidoreductase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4365 | - | 4e-09 | 23.20% (681) | hypothetical protein MSMEG_4365 |
| M. thermoresistible (build 8) | TH_2316 | - | 1e-10 | 25.45% (448) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5004 | - | 3e-85 | 34.41% (741) | hypothetical protein Mvan_5004 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK -MALDRYIGRVGALAVALGIG-----------SAVAAMPGQASAKPDADG
Mvan_5004|M.vanbaalenii_PYR-1 -MGYAQFVGRVGALAVALGVG-----------AATVALPGAAWAEPSDAA
MSMEG_4365|M.smegmatis_MC2_155 -------MGRIGALAVALGIG-----------AALTTG-TGVAWADDSGA
TH_2316|M.thermoresistible__bu -------VGRVGALAVALGVG-----------VAVATGGTAVAWAD-SGS
MAB_4293|M.abscessus_ATCC_1997 MSTATITLGIIGVIFSLVAWGSFFGGVVKMVRVILSGQPDRTRFRPFLPR
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK GSG----------MSETSVSGPARAGSGTDATSRSGKPAQSERSAATKPS
Mvan_5004|M.vanbaalenii_PYR-1 SSSSADNTKAEDTAEDSTAADDARPDDAVDDEVNSGEDHDEEAPQSEDGG
MSMEG_4365|M.smegmatis_MC2_155 SGGSTSSSSSASDS------SSSSAGGSSGSASG----------------
TH_2316|M.thermoresistible__bu TASSSSSQDSSSKSNPAQTRSSAKSGIGSGAGSGPGSRLDRPASRAPLGG
MAB_4293|M.abscessus_ATCC_1997 LKQLIVEVVAHTRMNKFRTVGWAHWLVMVGFLGGFPLYFESYGQTFNPEF
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK GGSDPPDRSPR---------------------------------EVAVTE
Mvan_5004|M.vanbaalenii_PYR-1 DGSDEDGGQPEKKTRGSYGSNRSEGELTGEDADEARADREDTDTEADVDV
MSMEG_4365|M.smegmatis_MC2_155 -GADAAKPRKPGVTKP--------------------------RIQPTGKI
TH_2316|M.thermoresistible__bu SGADSSATEQTGAVSSGTSAWRSAGM--------------PAGTSLTGRL
MAB_4293|M.abscessus_ATCC_1997 HWPIIGDTFLWHLWDELLGIGTVIGIVTLIIIRQLNHPRKPERLSRFGGS
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK ETTSGETADDEAEPVEETRDTVEIVPP----------------------T
Mvan_5004|M.vanbaalenii_PYR-1 VAPEAEPTADEADDEADGGDPAEVAEPGVADDVAVEQPAEVSDDAPEPVE
MSMEG_4365|M.smegmatis_MC2_155 AESLKKTAKAVEGAVADAHKRAQQSAA-------------------QQAE
TH_2316|M.thermoresistible__bu ADSVRDAVKPWRPATTRSTDPTPVETP-------------------AEAN
MAB_4293|M.abscessus_ATCC_1997 NFFAAYTIEMIVLVEGLGMVLVKSGKIATFHNSHPSSDFFTMNVAQLLPE
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK EPVTPASAVET---------EDEPDSPASSSLLWFLGAAGRRQTDQQAGP
Mvan_5004|M.vanbaalenii_PYR-1 SPISAPSSIVTALFAPKSWGDTAPADPVESPLLWTLLAFARRQFGQPRTE
MSMEG_4365|M.smegmatis_MC2_155 KRAERRAKLRAQIRPGKDTDKQAPEKQ---------AGNEAKNAAQPATK
TH_2316|M.thermoresistible__bu PEAQPDAHAGSPGESTAPETQPAPSSRGIRSMTAVLPDTESTNTESTNTE
MAB_4293|M.abscessus_ATCC_1997 SPIMVSVFAFIKMMSGGLFLLLVGRKLVWGVAWHRFAAFFNIYFKRNADG
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK DRDAASQTAAQAEAPIAATPVP----------------------------
Mvan_5004|M.vanbaalenii_PYR-1 IGDSGTPTGTTELVDPGAAAVPEPGEVTKGDPGIFTGTVRGQVKATDPDG
MSMEG_4365|M.smegmatis_MC2_155 TQRQLP--------AAPAVKVP----------------------------
TH_2316|M.thermoresistible__bu STVQHDDSERRSSLRSTAASVP----------------------------
MAB_4293|M.abscessus_ATCC_1997 SVALGAAKPMMSGGKVLEMESADPDVDAFGAGKVEDFSWKDLLDVTTCTE
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5004|M.vanbaalenii_PYR-1 GWLTYSGSTSTEKGTVTVTPWGTFRYTPSATARHAAAADGASTEAKTDTF
MSMEG_4365|M.smegmatis_MC2_155 --------------------------------------------------
TH_2316|M.thermoresistible__bu --------------------------------------------------
MAB_4293|M.abscessus_ATCC_1997 CGRCQSQCPAWNTGKPLSPKLLITSLRDHTYAKAPYLLAGEDKSKLSEAE
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5004|M.vanbaalenii_PYR-1 TVTVKDAAGNAVEVPVTVDILARNADPFGARARADNPSLTTGTAIIRVSA
MSMEG_4365|M.smegmatis_MC2_155 --------------------------------------------------
TH_2316|M.thermoresistible__bu --------------------------------------------------
MAB_4293|M.abscessus_ATCC_1997 IAEGERQLVGGE--------------------------------------
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5004|M.vanbaalenii_PYR-1 YDFDRDPLDITGPLSTGKGELVDNGDGTFTYTPTAAAREAASDPDAPDDA
MSMEG_4365|M.smegmatis_MC2_155 --------------------------------------------------
TH_2316|M.thermoresistible__bu --------------------------------------------------
MAB_4293|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK ------------------------------------GDASASPIIGVDGT
Mvan_5004|M.vanbaalenii_PYR-1 RIDTLTFTISDGHGGIRTATVDVIVAAYAESSASTPGRAAGPVLVSSNGT
MSMEG_4365|M.smegmatis_MC2_155 -------------------------------------DVITSVRARSDEV
TH_2316|M.thermoresistible__bu -------------------------------------ELSTPVRLADATA
MAB_4293|M.abscessus_ATCC_1997 -------------------------GAVIDHEVLWSCTTCGACVEQCPVD
Mb1842c|M.bovis_AF2122/97 --------------------------------------------------
Rv1812c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1994|M.gilvum_PYR-GCK VYQVTRTSSAGG--QFSTVTILDNTGQIVSSGEVRG----TVMDAVARPD
Mvan_5004|M.vanbaalenii_PYR-1 IYQVTYDLDSTNNPIRTRVSILDEDGQVLKTTDVIPGYPVEQALPVVRPD
MSMEG_4365|M.smegmatis_MC2_155 AREITTGVTGIRTAIENAAPRIALPDLRPTTNL-------ARPVTPVALK
TH_2316|M.thermoresistible__bu VRALAERLTERRSVTLESVR--AFTDVAATTALTSAASVAAATVEPVALI
MAB_4293|M.abscessus_ATCC_1997 IEHVDHIIDMRRYQVLIESEFPGELAGLFKNLENKGNPWGQNAKDRLNWI
Mb1842c|M.bovis_AF2122/97 ----------------------------------MTRVVVIGSGFAGLWA
Rv1812c|M.tuberculosis_H37Rv ----------------------------------MTRVVVIGSGFAGLWA
Mflv_1994|M.gilvum_PYR-GCK GTLVVFSRRFIS--TTVSTVSSDGTVTPIANLLGTPLNPPRVGPDGALYF
Mvan_5004|M.vanbaalenii_PYR-1 GSLLVTTYKASSNTSTVSIVDGQGEVKTLGTVIGQPSAPMTVAPNGAVFF
MSMEG_4365|M.smegmatis_MC2_155 AAQPVQ---VSPPRPTITGLVRGLLTAAAGFSPRAATAPVAPAPSPAAWA
TH_2316|M.thermoresistible__bu TAAPEATPAVDAEQPAAPRLVTGLL-AAVGLAPFTANSPLAPTQSTPLLA
MAB_4293|M.abscessus_ATCC_1997 EEVDFDVPVFGQDVDSFADFEYLFWVGCAGAYEDRAKKTTKAVAELLALA
.
Mb1842c|M.bovis_AF2122/97 A-----LGAARRLDELAVPAGT----------------VDVMVVSNKPFH
Rv1812c|M.tuberculosis_H37Rv A-----LGAARRLDELAVLAGT----------------VDVMVVSNKPFH
Mflv_1994|M.gilvum_PYR-GCK G-----AGLPDLLSPVGANVGYRTYRISPRNTVRAFAFDTDVAIADDGTA
Mvan_5004|M.vanbaalenii_PYR-1 KTRQFASGSGDRLVRVSATGGLRVYQLG--------VAADSPSVAPDGSV
MSMEG_4365|M.smegmatis_MC2_155 L-----LAFARREFERAFTPGAATALAAP--------AITTSAVTSAAVV
TH_2316|M.thermoresistible__bu L-----LGWARRETDRAYTQQARSLVTA-----------RPAVVSTAAVV
MAB_4293|M.abscessus_ATCC_1997 GTKFLVLGADETCTGDSARRAGNEFLFQQLAMQN-----IELLNSVFEGV
. . :
Mb1842c|M.bovis_AF2122/97 DIRVRNYEADLSACRIPLG----------------DVLGPAGVAHVTAEV
Rv1812c|M.tuberculosis_H37Rv DIRVRNYEADLSACRIPLG----------------DVLGPAGVAHVTAEV
Mflv_1994|M.gilvum_PYR-GCK YLVSRRYGISVLRVIPTAGLGRTTLLPLGTDPS-APVLGPDGTAHVTAGV
Mvan_5004|M.vanbaalenii_PYR-1 YIVSRSFGVTSVLAVGPGGNSRRVSLPLGADTVNDVVIGQDGRGYLTVER
MSMEG_4365|M.smegmatis_MC2_155 NPLPGRLD--SVPVGWVTGVSNPSGNWPQTNNTAGFDIWGTDLGIMWDGG
TH_2316|M.thermoresistible__bu DPLPADLIGETERIGRITGPG-------ITDGPNRLNIYGTDLGIMWDMG
MAB_4293|M.abscessus_ATCC_1997 EQKQRKIVVTCAHCFNALGNEYPQVGGDYQVVHHTQLLNRLVRDKKLVPV
* :
Mb1842c|M.bovis_AF2122/97 TAIDADGRRVTTSTGASYS---YDRLVLASGSHVVKPALPGLAEFGFDVD
Rv1812c|M.tuberculosis_H37Rv TAIDADGRRVTTSTGASYS---YDRLVLASGSHVVKPALPGLAEFGFDVD
Mflv_1994|M.gilvum_PYR-GCK TWFGSPQTRVYTATGDSIAAHTVAG--LPAGAVVG-ADGVYLATFVEPGS
Mvan_5004|M.vanbaalenii_PYR-1 NVFGTKTTRVYTFTGTSNTVREIPG--TPDGAKVITADGVYQYTYDES--
MSMEG_4365|M.smegmatis_MC2_155 EWNG--QRFVHTAFGDTFSGPNMSGWWRSNVLLISTDNVLSDGLG--LVQ
TH_2316|M.thermoresistible__bu EWRG--QRAVHVVFGDTFSGPNMSGDWISNAIMISTSRTLYENPAGFLEQ
MAB_4293|M.abscessus_ATCC_1997 APVSQDVTYHDPCYLGRHNKVYTAPRELIGASGAAITEMPRHGERSMCCG
.
Mb1842c|M.bovis_AF2122/97 TYDGAVRLQQHLQGLAGGPLTSAAATVVVVGAGLTG--------------
Rv1812c|M.tuberculosis_H37Rv TYDGAVRLQQHLQGLAGGPLTSAAATVVVVGAGLTG--------------
Mflv_1994|M.gilvum_PYR-GCK EGPGTSYVSRLTAGGVGTSGLIEG-RIAEFQVTAEG--------------
Mvan_5004|M.vanbaalenii_PYR-1 --TGKSYISRITADTIETSDPIDGRVINPISVTPDG--------------
MSMEG_4365|M.smegmatis_MC2_155 TGTAYQFIPASGRNLFGLFGGSEVTVIPTAGVQIDG--------------
TH_2316|M.thermoresistible__bu TGPAFQFIP--GGQLLSWFFPAEVTVIPTAGVHANG--------------
MAB_4293|M.abscessus_ATCC_1997 AGGARMWMEEQLGKRINIDRVDEALSTPASKIATGCPFCRVMLTDGVTAR
. :
Mb1842c|M.bovis_AF2122/97 IETACELPGRLHALFARGDGVTPRVVLIDHN--PFVGSDMGLSARPVIEQ
Rv1812c|M.tuberculosis_H37Rv IETACELPGRLHALFARGDGVTPRVVLIDHN--PFVGSDMGLSARPVIEQ
Mflv_1994|M.gilvum_PYR-GCK TVFAPIDG-AGAETPVAVIDRDGTVHTVLLP-GTLVVGRRAIR-----DG
Mvan_5004|M.vanbaalenii_PYR-1 TVYVAVRNSATGTDSVAIISTSGEVTTVDIP-GTIFPVLPSVSPAVTGDD
MSMEG_4365|M.smegmatis_MC2_155 TQYVNYMSVRSWDTPGRWTTNFSAISVYDPSTDTWVLAPSTIRSAGWFRS
TH_2316|M.thermoresistible__bu TQYVNYMSVRQWGVPGSWTTNFSAISRYDQATDTWVTVPSSVRSAGWLRS
MAB_4293|M.abscessus_ATCC_1997 DDSAAVEVVDVAQLLLESVGRTEDIRKALPAKGTAAAAAAERAATKTAEP
. : .
Mb1842c|M.bovis_AF2122/97 ALLDNGVETRTGVSVAAVSPGGVTL-SSGERLAAATVVWCAGMRASRLTE
Rv1812c|M.tuberculosis_H37Rv ALLDNGVETRTGVSVAAVSPGGVTL-SSGERLAAATVVWCAGMRASRLTE
Mflv_1994|M.gilvum_PYR-GCK GTYNVEDVGYVNYATGGREYVAVLS-ADGT-VARTIELPEGATGGS-VFY
Mvan_5004|M.vanbaalenii_PYR-1 ANPNIGDNGYVAYQSGGVSYLAVVN-PDGT-IARTVTLPAGAVVATPVDF
MSMEG_4365|M.smegmatis_MC2_155 STLYVPGSQNFQQAAYVLQPEDQVG-EDGIRYLYAFGTPSGRAGSAYLSR
TH_2316|M.thermoresistible__bu STPYRAGDQNFQQMAYVLQPVDQVG-EDGVRYVYAFGTPSGRQGSVHLSR
MAB_4293|M.abscessus_ATCC_1997 EPVVAEEEAPATTKAEAAAPAAEAKPVSGLGMATGAKRPGAKKTAAENST
.* . .
Mb1842c|M.bovis_AF2122/97 QLPVARDRLGRLQVDDYLRVIGVPAMFAAGDVAAARMDDEHLSVMSCQHG
Rv1812c|M.tuberculosis_H37Rv QLPVARDRLGRLQVDDYLRVIGVPAMFAAGDVAAARMDDEHLSVMSCQHG
Mflv_1994|M.gilvum_PYR-GCK SPDGAAFEMLEYRDAQGQTTFRQILALASTTYTDKVPGGRFTYVRDTVVF
Mvan_5004|M.vanbaalenii_PYR-1 GPDGAAYQVIETRDEQGRVTSRAVLALSTDTVTPMLPGAPLQPNYPSIQF
MSMEG_4365|M.smegmatis_MC2_155 VPEDAVTDLSKYEYWDGNRWVTGNAAVAAPVIGDSTRSTGLFGFIVDWAN
TH_2316|M.thermoresistible__bu VPEEHITDVSKYEYWDGNAWVS-SAAHAAPIIGESTRSAGLFGFIIDWAN
MAB_4293|M.abscessus_ATCC_1997 AEASTATPTAEAAPAAPVKGLGMAAGAKRPGAKKATAETSAATPPAASAS
.
Mb1842c|M.bovis_AF2122/97 RP--MGRYAG------------------------CNVINDLFDQPLLALR
Rv1812c|M.tuberculosis_H37Rv RP--MGRYAG------------------------CNVINDLFDQPLLALR
Mflv_1994|M.gilvum_PYR-GCK GPDGVGYLLTGSVSTQIGASQDLDVLGFNAAGETVSRAGGLTNPVTTYEN
Mvan_5004|M.vanbaalenii_PYR-1 GPDGSGVLITVETGQSP---FEYHFLRFDQDGATIATADLSGFLQSAQQD
MSMEG_4365|M.smegmatis_MC2_155 DPNVLGGYLGGLVGAKTGGNVSELSVQYNEYLGKYVVLYGDG-NNNIQMR
TH_2316|M.thermoresistible__bu DPNVFGGYLGGLFGAKTGGNVGEMSVQYNEYLGKYVILYGNGRTNNIEMR
MAB_4293|M.abscessus_ATCC_1997 PPASQAPAAAAAPAPPVKGLGMAAGAKRPGAKKTAPAAPAAPAPEAAAVP
* .
Mb1842c|M.bovis_AF2122/97 IPWYVTVLDLGSAGAVYTEGWER-------KVVSQGAPAKTTKQSINTRR
Rv1812c|M.tuberculosis_H37Rv IPWYVTVLDLGSAGAVYTEGWER-------KVVSQGAPAKTTKQSINTRR
Mflv_1994|M.gilvum_PYR-GCK NPYRTV-LAFGRDGTAYVTLYFTPDDPGVYAVTATGARKVVDLEYVQYRQ
Mvan_5004|M.vanbaalenii_PYR-1 YVFWQEGVVFGPDGTPYATLTG--ADQGVWALTSTGPVKVLELDLGQGEL
MSMEG_4365|M.smegmatis_MC2_155 IADRPEGPWSDPIELASSADYPGLYAPMIHPWSGTGMLEDDNGDPDVNNL
TH_2316|M.thermoresistible__bu IADSPEGPWSDPIILATPQQYPGLYAPMIHPWSGTGYLLDENGDPDPESL
MAB_4293|M.abscessus_ATCC_1997 SESEAPTATETSAVAEPPVKGLGIAAGARRPGAKKAAPATESAPEPAQPA
.
Mb1842c|M.bovis_AF2122/97 IYPPLNGSRADLLAAAAPRVQPRP--------------------
Rv1812c|M.tuberculosis_H37Rv IYPPLNGSRADLLAAAAPRVQPRP--------------------
Mflv_1994|M.gilvum_PYR-GCK GFPPTFGLDGTGYVTSAELLSGDADGIALVTVFSPVTDL-----
Mvan_5004|M.vanbaalenii_PYR-1 VEPVKFGPDGTPYVTVSERVDG--SYVTTVHTFTPVTML-----
MSMEG_4365|M.smegmatis_MC2_155 YWNMSLWGDYNVVLMQTDLSGLKTVAV-----------------
TH_2316|M.thermoresistible__bu YWNMSMWGPYNVYLMQTDLSSLVEV-------------------
MAB_4293|M.abscessus_ATCC_1997 TAPEPTEPATSAPKDEAPETSSTPEPPVKGLGIAPGARRPGRRN
: