For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNDALGSVRQKLQQVSESGRALKRLIDTGIIDFSDLPASVRTAKLSRVYGPQATMAIQGGRKYADLPAIV DERGTLTYKQVDDQSWALAHGLRRLGVSAGSVVGVLCRDHRGLVITMAACGKLGARMVLMNTGFAKPQFA EVCKRENVAVVLHDSEFLGLLEALPADMPRVLTWVDDGTEVPPGVPTLDDIVTANSTEPLPAPDKSGGSV ILTSGTTGLPKGAPRDSVSPLATAQIIDRIPFPHKGTMVIVSPIFHSTGWATYTVGAAFGNKIVTSRRFK AEKTLELIATHKADMLVAVPTMLHRMVELGPDVIAKYDTSSLKVILIAGSALSPELSNRVQDTFGDVLYN MYGSTECAIASVATPAELRAAPGTAGRAPVTCEVVLYDENDQRIHGTNRRGRIFVRNGAPFSGHTDGRSK QIIDGFMSSGDMGHFTDDGLLFVDGRDDDMIVSGGENVFPQEVEQLLEERPDVAEVAVVGVDDVEFGKRL RAFIVTEPGAAREPEEIKRHVKENLARHKVPRDVVFVDELPRNATGKLLRRVLVEMDVES
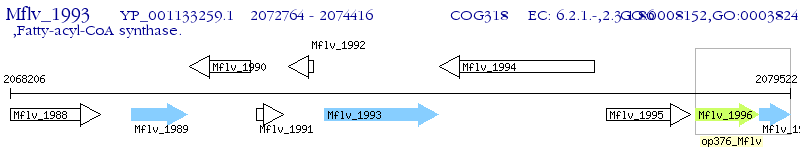
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1993 | - | - | 100% (550) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_0435 | - | e-110 | 41.92% (520) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_0341 | - | e-108 | 41.67% (492) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0276 | fadD2 | 1e-114 | 41.26% (555) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv0270 | fadD2 | 1e-114 | 41.26% (555) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_02546 | fadD2 | 1e-113 | 40.66% (546) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4340c | - | 1e-115 | 40.51% (548) | acyl-CoA synthetase |
| M. marinum M | MMAR_0528 | fadD2 | 1e-112 | 39.96% (548) | fatty-acid-CoA ligase FadD2 |
| M. avium 104 | MAV_4891 | - | 1e-110 | 40.80% (549) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_0304 | - | 1e-110 | 42.40% (526) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_4533 | - | 1e-112 | 39.49% (547) | PUTATIVE acyl-CoA synthase |
| M. ulcerans Agy99 | MUL_1191 | fadD2 | 1e-112 | 39.96% (548) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_2344 | - | 1e-136 | 48.86% (528) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0528|M.marinum_M MPNLSDLPG-----VAKIQ-----QYVDRGAAELHYVRKIIESGAFRLES
MUL_1191|M.ulcerans_Agy99 MPNLSDLPG-----VAKIQ-----QYVDRGAAELHYVRKIIESGAFRLES
MAV_4891|M.avium_104 MPNLLGLPGQATKAVAKVQ-----QYVERGSAELHYLRRIIESGAFRLEP
Mb0276|M.bovis_AF2122/97 MPNLTDLPG---QAVSKLQKS-IGQYVARGTAELHYLRKIIESGAIGLEP
Rv0270|M.tuberculosis_H37Rv MPNLTDLPG---QAVSKLQKS-IGQYVARGTAELHYLRKIIESGAIGLEP
MLBr_02546|M.leprae_Br4923 --------------MTKLR-----HYLERGAAEVHYLRSIFESGALGFEP
TH_4533|M.thermoresistible__bu MAKLTDLPR---LAADKLPLSRLQLYAERGAAELHYLRKMLEAGALKIES
MAB_4340c|M.abscessus_ATCC_199 MSITAERPL-----FSKLQ-----TLAKRGGAELHYLRKVIEAGMLKLDP
Mflv_1993|M.gilvum_PYR-GCK ----------MNDALGSVR-----QKLQQVSESGRALKRLIDTGIIDFSD
Mvan_2344|M.vanbaalenii_PYR-1 ---------------MSAS-----GRLLHTVTLARASVVLCKRGMVDPLR
MSMEG_0304|M.smegmatis_MC2_155 --------------MGLLD-----RITDPLIDTVGMVTTMRRAGLIAPLR
: * .
MMAR_0528|M.marinum_M PLNYAAMATEIAKWGEFGMLPSFNARR-HPHRAACIDEEGEFTYQQLDEA
MUL_1191|M.ulcerans_Agy99 PLNYAAMATEIAKWGEFGMLPSFNARR-HPHRAACIDEEGEFTYQQLDEA
MAV_4891|M.avium_104 PQNYAAMAADIYKWGEFGMLPSLNARR-TPDRAAVIDEEGELSYAELDRA
Mb0276|M.bovis_AF2122/97 PLNYAALAADIRKWGEVGMLPSHNARR-APNRAAVIDEEGTLTFSELDEA
Rv0270|M.tuberculosis_H37Rv PLNYAALAADIRKWGEVGMLPSHNARR-APNRAAVIDEEGTLTFSELDEA
MLBr_02546|M.leprae_Br4923 PLTYAAMAVAMRKWGDLAMLPAMPARR-SPHRAALIDEEGTLTYKELDRA
TH_4533|M.thermoresistible__bu PTRIAALLADIRRWGEIGMIPALNARR-WPDRVAVIDDEGEMTFAELNDA
MAB_4340c|M.abscessus_ATCC_199 PNVLAGLVRDFYRFGEIGAVPAFGAHR-SPSRTAIIDDEGSITYAELNDA
Mflv_1993|M.gilvum_PYR-GCK LPASVRTAKLSRVYGPQATMAIQGGRK-YADLPAIVDERGTLTYKQVDDQ
Mvan_2344|M.vanbaalenii_PYR-1 PRDALRASRAVRRYGAFGGLASFTAAQ-YRHWPALTDDTETLTFVELDQQ
MSMEG_0304|M.smegmatis_MC2_155 PDKYLRIVAAMARENMSVTSGFASAAQRCPDRPGLVDELGTLTWREIDQQ
. . : . *: ::: :::
MMAR_0528|M.marinum_M AHAVANALLAKGVSAGDGVAILARNHRWFLIANYGAARAGARIILLNSEF
MUL_1191|M.ulcerans_Agy99 GHAVANALLAKGVSAGDGVAILARNHRWFLIANYGAARAGARIILLNSEF
MAV_4891|M.avium_104 AHAVANGLIAKGVKAGDGVAILARNHRWFLIANYGAARVGARIILLNSEF
Mb0276|M.bovis_AF2122/97 AHAVANGLLAKGVRAGDGVAILARNHRWFVIANYGAARVGARIILLNSEF
Rv0270|M.tuberculosis_H37Rv AHAVANGLLAKGVRAGDGVAILARNHRWFVIANYGAARVGARIILLNSEF
MLBr_02546|M.leprae_Br4923 AHALANGLIAKGVRGGDGIAILARNHRWFVIANYGCARVGARIILLNSEF
TH_4533|M.thermoresistible__bu AHAVANALLAKGVRGGDGVAILARNHRWFLVSMYGAARVGARIILLNTEF
MAB_4340c|M.abscessus_ATCC_199 VNALAHGLNRLGIKGGDGVAILARNHRWFIIANYAAHRAGARVILLNTDF
Mflv_1993|M.gilvum_PYR-GCK SWALAHGLRRLGVSAGSVVGVLCRDHRGLVITMAACGKLGARMVLMNTGF
Mvan_2344|M.vanbaalenii_PYR-1 SNARAQAIP-AAAGGPAVVGLLRRSGVELVLDLAAANKAGVRTVLLNTGF
MSMEG_0304|M.smegmatis_MC2_155 ADALAAGLQALSGGAPRVLGIMARNHRGFVLSLIAANRIGADVLLLNTSF
* * .: . . :.:: *. ::: .. : *. :*:*: *
MMAR_0528|M.marinum_M SGPQIKEVSEREGAKLIIYDDEYTGAVSKAEPPLGKLRALGVNPDSEEDS
MUL_1191|M.ulcerans_Agy99 SGPQIKEVSEREGAKLIIYDDEYTGAVSKAEPPLGKLRALGVNPDSEEDS
MAV_4891|M.avium_104 SGPQIKEVSEREGAKVIIYDDEYTKAVSKAEPPLGKLRALGSNPDADEPS
Mb0276|M.bovis_AF2122/97 SGPQIKEVSDREGAKVIIYDDEYTKAVSLAQPPLGKLRALGVNPDDDKPS
Rv0270|M.tuberculosis_H37Rv SGPQIKEVSDREGAKVIIYDDEYTKAVSLAQPPLGKLRALGVNPDDDKPS
MLBr_02546|M.leprae_Br4923 SGPQIKEVSEREGAKVIIYDDEYTKVVSKAEPALGKLRALGVNPDEEEPP
TH_4533|M.thermoresistible__bu SGPQICEVSDREGAKLIIYDDEYTAAVSGVEPELGKLRALGHNPDSDEPS
MAB_4340c|M.abscessus_ATCC_199 SGPQTKEVAEREGARVLIYDAEYAEFLDGYSPELGRIMALPTNPDNPDQP
Mflv_1993|M.gilvum_PYR-GCK AKPQFAEVCKRENVAVVLHDSEFLGLLEALPADMPRVLTW---VDDGTEV
Mvan_2344|M.vanbaalenii_PYR-1 AAPQLADVCLREGVTTVVADDEFAPLLRDLDPDVVRLT--------GTQM
MSMEG_0304|M.smegmatis_MC2_155 SGPALAEVVSREKVDAVIYDEEFTDTVDRALAGAPEAPTR---IVAWTDT
: * :* ** . :: * *: : . .
MMAR_0528|M.marinum_M GSTDETLAELIARSSTAPAPKAGKHASIIILTSGTTGTPKGANRSTPPTL
MUL_1191|M.ulcerans_Agy99 GSTDETLAELIARSSTAPAPKAGKHASIIILTSGTTGTPKGANRSTPPTL
MAV_4891|M.avium_104 GSTDETLAELIERSSSEPAPKADRHASIIILTSGTTGTPKGANRSTPPTL
Mb0276|M.bovis_AF2122/97 GSSDETLAELIAHSSTAPAPKASRRASIIILTSGTTGTPKGANRNTPPTL
Rv0270|M.tuberculosis_H37Rv GSSDETLAELIAHSSTAPAPKASRRASIIILTSGTTGTPKGANRNTPPTL
MLBr_02546|M.leprae_Br4923 GSTDETLAELIARSSSAPAPKVRKRASIIILTSGTTGTPKGANRNTPASL
TH_4533|M.thermoresistible__bu GSTDETLADIIERVGAKPAPKATRHSSIIILTSGTTGTPKGATRKAPPSL
MAB_4340c|M.abscessus_ATCC_199 ASTDETIASVIKRNSTSPAPRPSKYSSLVILTSGTTGTPKGAPRKLALTL
Mflv_1993|M.gilvum_PYR-GCK PPGVPTLDDIVTANSTEPLPAPDKSGGSVILTSGTTGLPKGAPRDSVSPL
Mvan_2344|M.vanbaalenii_PYR-1 AS-------RAATKSTAIPVVPRRPGGLVLLTSGTTGIPKGAPRDRLNPL
MSMEG_0304|M.smegmatis_MC2_155 AAHDVTVAALTEKFAGRQPQRGGQKSKVILLTSGTTGTPKGAKHSGGGPE
. . : . ::******* **** :. .
MMAR_0528|M.marinum_M APVGGILSHVPFKANEVTSLPAPMFHALGYLHATIAMFLGSTLVLRRKFK
MUL_1191|M.ulcerans_Agy99 APVGGILSHVPFKANEVTSLPAPMFHALGYLHATIAMFLGSTLVLRRKFK
MAV_4891|M.avium_104 APVGGILSHVPFKAGEVTSLPSPMFHALGYLHATIAMFLGSTLVLRRKFK
Mb0276|M.bovis_AF2122/97 APIGGILSHVPFKAGEVTLLPSPMFHALGYMHAALAMFLGSTLVLRRRFK
Rv0270|M.tuberculosis_H37Rv APIGGILSHVPFKAGEVTLLPSPMFHALGYMHAALAMFLGSTLVLRRRFK
MLBr_02546|M.leprae_Br4923 APLGGILSKVPFRAHEVTLLPAPMFHALGYLHATLAMFLGSTLVLRRRFK
TH_4533|M.thermoresistible__bu APIGGVLSHVPFKSGEVTSLPAPMFHALGYLHATIAMMLGNTLLLRRRFK
MAB_4340c|M.abscessus_ATCC_199 APVGGMFSHVPFRSGEVTSVPAPMFHALGYLHQSLALTLGCTLILRRKFK
Mflv_1993|M.gilvum_PYR-GCK ATA-QIIDRIPFPHKGTMVIVSPIFHSTGWATYTVGAAFGNKIVTSRRFK
Mvan_2344|M.vanbaalenii_PYR-1 QSA-QLLDRIPWPRNSAYVVAAPLFHATGLATCTVGLALGNHVVLHRRFD
MSMEG_0304|M.smegmatis_MC2_155 ILK-AILDRTPWRAEEPVVIVAPMFHAWGFSQLAFAASMACTIITRRKFD
::.: *: : :*:**: * :.. :. :: *:*.
MMAR_0528|M.marinum_M PQLVLEDIEKHKVTAMVVVPVMLSRLLDAVEKIDKKPDLSSLKIVFVSGS
MUL_1191|M.ulcerans_Agy99 PQLVLEDIEKHKVTAMVVVPVMLSRLLDAVEKIDKKPDLSSLKIVFVSGS
MAV_4891|M.avium_104 PPLVLQDIEKYRPTAMVVVPVMLSRILDTLEKMDKKPDLSSLRIVFVSGS
Mb0276|M.bovis_AF2122/97 PALVLEDIEKHKATSMVVVPVMLSRILDQLEKTEPKPDLSSLKIVFVSGS
Rv0270|M.tuberculosis_H37Rv PALVLEDIEKHKATSMVVVPVMLSRILDQLEKTEPKPDLSSLKIVFVSGS
MLBr_02546|M.leprae_Br4923 PATVLEDIEKHQATAMVVVPVMLSRILDTLENLETKSDLSSLRVVFVSGS
TH_4533|M.thermoresistible__bu PATVLTDIEKHNVSAVIVVPVMLSRMLDEWEKMRPKPDLSSLRIVFVSGS
MAB_4340c|M.abscessus_ATCC_199 PENVLADVEKFKVTAIVVVPVMLNRLLNALDELSVKPDLSSLRIVFVSGS
Mflv_1993|M.gilvum_PYR-GCK AEKTLELIATHKADMLVAVPTMLHRMVELGPDVIAKYDTSSLKVILIAGS
Mvan_2344|M.vanbaalenii_PYR-1 ARETLEAVQKHHARALILVPTMLSRILDLPPAALNAFDTSSLSVVFVAGS
MSMEG_0304|M.smegmatis_MC2_155 PEATLELVDRHRATGLCVVPVMFDRIMELPEDVLNRYDGRSLRFAAASGS
. .* : .. : **.*: *::: * ** . :**
MMAR_0528|M.marinum_M QLGAELASRALKDLGPVIYNMYGSTEIAFATIAGPEDLQRNAATVGPVVK
MUL_1191|M.ulcerans_Agy99 QLGAELASRALKDLGPVIYNMYGSTEIAFATIAGPEDLQRNAATVGPVVK
MAV_4891|M.avium_104 QLGAELAARALKDIGPVIYNMYGSTEIAFATIAGPKDLERNAATVGPVVK
Mb0276|M.bovis_AF2122/97 QLGAELATRALGDLGPVIYNMYGSTEVAFATIAGPKDLQFNPSTVGPVVK
Rv0270|M.tuberculosis_H37Rv QLGAELATRALGDLGPVIYNMYGSTEVAFATIAGPKDLQFNPSTVGPVVK
MLBr_02546|M.leprae_Br4923 QLGAELATRALEELGPVIYNMYGSTEIAFATIAGPKDLQINSATVGPVVK
TH_4533|M.thermoresistible__bu QLGAELASRAMRDLGPVIYNLYGSTEIAFATIARPQDLSINPATVGPVVK
MAB_4340c|M.abscessus_ATCC_199 QLGGELASRAMDTLGPVIYNLYGSTEVAFATIARPQDLAINPSTVGPVVK
Mflv_1993|M.gilvum_PYR-GCK ALSPELSNRVQDTFGDVLYNMYGSTECAIASVATPAELRAAPGTAGRAPV
Mvan_2344|M.vanbaalenii_PYR-1 SLSPELYRRAAAHFGEVLYNLYGSTEVATAAVATPAELRRAPGTVGRPPI
MSMEG_0304|M.smegmatis_MC2_155 RMRPDVVIAFMDRFGDVIYNNYNATEAGMIATATPADLRAAPDTAGRPAE
: :: :* *:** *.:** . : * * :* . *.*
MMAR_0528|M.marinum_M GVKVKILDD-NGNEVSQGQVGRIFVGNAFPFEGYTGGGHKQIIDGLMSSG
MUL_1191|M.ulcerans_Agy99 GVKVKILDD-NGNEVSQGQVGRIFVGNAFPFEGYTGGGHKQIIDGLMSSG
MAV_4891|M.avium_104 GVKVKIFDD-NGKELPQGEVGRIFVGNTFPFAGYTGGGNKQIIDGLLSSG
Mb0276|M.bovis_AF2122/97 GVTVKILDE-NGNEVPQGAVGRIFVGNAFPFEGYTGGGGKQIIDGLLSSG
Rv0270|M.tuberculosis_H37Rv GVTVKILDE-NGNEVPQGAVGRIFVGNAFPFEGYTGGGGKQIIDGLLSSG
MLBr_02546|M.leprae_Br4923 GVKVKILDD-SGKEVPRGQVGRIFVGNAFPFEGYTGGGGKQIIDGLLSSG
TH_4533|M.thermoresistible__bu GVTVKLFDE-NGKEVPRGEVGRIFVRNTFPFEGYTGGGHKQIIDGLMSSG
MAB_4340c|M.abscessus_ATCC_199 GATVKILDD-EGKPVSQGTVGRIFVSNAIPFDGYTGGGHKQIIDGLMSSG
Mflv_1993|M.gilvum_PYR-GCK TCEVVLYDENDQRIHGTNRRGRIFVRNGAPFSGHTDGRSKQIIDGFMSSG
Mvan_2344|M.vanbaalenii_PYR-1 GCTIAAYDSRDRRITRAGEVGTIYVSSGLSFGGYTGGGSKKTVDGLLSSG
MSMEG_0304|M.smegmatis_MC2_155 GTEIRILDG-DLRDLPTGEVGTIYVRNSTQFDGYTSGTTKDFHEGFMCSG
: * . . . * *:* . * *:*.* *. :*::.**
MMAR_0528|M.marinum_M DVGYFDEHGLLYVSGRDDEMIVSGGENVFPAEVEDLISGHPDVVEATALG
MUL_1191|M.ulcerans_Agy99 DVGYFDEHGLLYVSGRDDEMIVSGGENVFPAEVEALISGHPDVVEATALG
MAV_4891|M.avium_104 DVGYFDEHGLLYVSGRDDEMIVSGGENVFPAEVEDLISGHPDVVEATAIG
Mb0276|M.bovis_AF2122/97 DVGYFDERGLLYVSGRDDEMIVSGGENVFPAEVEDLISGHPDVVEAAAIG
Rv0270|M.tuberculosis_H37Rv DVGYFDERGLLYVSGRDDEMIVSGGENVFPAEVEDLISGHPDVVEAAAIG
MLBr_02546|M.leprae_Br4923 DVGYFDEHDLLYISGRDDEMIVSGGENVFPAEVEDLISGHPEVVEATAIG
TH_4533|M.thermoresistible__bu DVGYFDEHGLLYVSGRDDEMIVSGGENVFPAEIEDLLSGHPEIVEATAIG
MAB_4340c|M.abscessus_ATCC_199 DVGYFDEHGLLYVSGRDDEMIVSGGENVFPAEVEDLISGHPEVIEATALG
Mflv_1993|M.gilvum_PYR-GCK DMGHFTDDGLLFVDGRDDDMIVSGGENVFPQEVEQLLEERPDVAEVAVVG
Mvan_2344|M.vanbaalenii_PYR-1 DTGHFDDDGLWFIDGREDDMVISGGENVYPLEIENLLVELPGVREAAVVG
MSMEG_0304|M.smegmatis_MC2_155 DVGRLDDAGRLFVVGRDDEMIVSGGENVYPIEVEKVLTTHPAVAEAAVIG
* * : : . :: **:*:*::******:* *:* :: * : *.:.:*
MMAR_0528|M.marinum_M VDDKEWGARLRAFVVKKPDTDLDEETVKHYVRDHLARYKVPREVIFLEEL
MUL_1191|M.ulcerans_Agy99 VDDKEWGARLRAFVVKKPDTDLDEETVKHYVRDHLARYKVPREVIFLEEL
MAV_4891|M.avium_104 VEDKEWGHRLRAFVVKKEGADLDEDTIKHYVRDHLARYKVPREVIFLDEL
Mb0276|M.bovis_AF2122/97 VDDKEFGARLRAFVVKKPGADLDEDTIKQYVRDHLARYKVPREVIFLDEL
Rv0270|M.tuberculosis_H37Rv VDDKEFGARLRAFVVKKPGADLDEDTIKQYVRDHLARYKVPREVIFLDEL
MLBr_02546|M.leprae_Br4923 VDDKEWGARLRAFVVKKQDATIDEDAIKAYVRDHLARYKVPREVIFLEEL
TH_4533|M.thermoresistible__bu VEDKEWGQRLRAFVVKAEGSDLDEDEVKRYVREHLARYKVPRDVIFLDEL
MAB_4340c|M.abscessus_ATCC_199 VDDPEWGARLRAFVVRHPDSTVDADGIRAYVRDNLARYKVPRDVVFLDVL
Mflv_1993|M.gilvum_PYR-GCK VDDVEFGKRLRAFIVTEPGAAREPEEIKRHVKENLARHKVPRDVVFVDEL
Mvan_2344|M.vanbaalenii_PYR-1 VRDDDFGMCLRAFVVPAAGADIDADQIRHAVRSRLARHKVPRDVLFVDAL
MSMEG_0304|M.smegmatis_MC2_155 VDDEQFGQRLAAFVVLSGDATVD--DLKSHVRDNLANYKVPRDITILDEL
* * ::* * **:* .: : :: *:..**.:****:: ::: *
MMAR_0528|M.marinum_M PRNPTGKILKRELREMEIE-
MUL_1191|M.ulcerans_Agy99 PRNPTGKILKRELREMEIE-
MAV_4891|M.avium_104 PRNPTGKILKRELREMEV--
Mb0276|M.bovis_AF2122/97 PRNPTGKVLKRELRKL----
Rv0270|M.tuberculosis_H37Rv PRNPTGKVLKRELRKL----
MLBr_02546|M.leprae_Br4923 PRNPTGKVLKRELRDFEVH-
TH_4533|M.thermoresistible__bu PRNPTGKILKRELRELEVS-
MAB_4340c|M.abscessus_ATCC_199 PRNPSGKILKRELRELEV--
Mflv_1993|M.gilvum_PYR-GCK PRNATGKLLRRVLVEMDVES
Mvan_2344|M.vanbaalenii_PYR-1 PRNETGKVLKRDLARMEQR-
MSMEG_0304|M.smegmatis_MC2_155 PRNSTGKIARRELQEKVNG-
*** :**: :* *