For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RCKPGRPDLDAYATYFDLPAATPASPVTVSWAGVTTLLINDGTSALMTDGFFSRPGLLRVATRRPLASSD SRIDAALQRLGVTRLDGVTPVHTHFDHAMDSAAVAGRTGARLIGGSSAAHIGRGHGLDRVEVVTSGDATA VGAFDVTLIESHHCPPDRFPGAITAPVAPRASVSAYRCGEAWSTLVRHRGTGRGLLIVGSAGFVPGALAG RHADVVYLGVGQLGLQPHDYLAAYWTETVRTVGARRVVLIHWDDFFRPLDKPLRALPFVADNLDVSMRIL ARLAREDGVDLHLPTLWERADPWR*
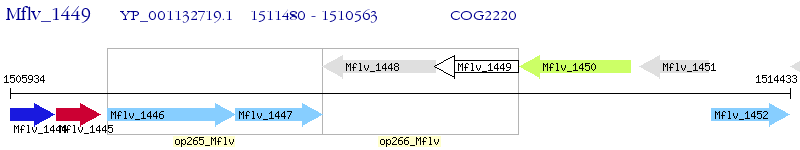
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1449 | - | - | 100% (305) | hypothetical protein Mflv_1449 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3608 | - | 1e-108 | 64.93% (288) | hypothetical protein Mb3608 |
| M. tuberculosis H37Rv | Rv3577 | - | 1e-108 | 64.93% (288) | hypothetical protein Rv3577 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_5077 | - | 1e-108 | 61.44% (306) | hypothetical protein MMAR_5077 |
| M. avium 104 | MAV_0579 | - | 1e-114 | 64.05% (306) | hypothetical protein MAV_0579 |
| M. smegmatis MC2 155 | MSMEG_6071 | - | 1e-125 | 71.90% (306) | metallo-beta-lactamase superfamily protein |
| M. thermoresistible (build 8) | TH_1750 | - | 1e-118 | 67.43% (307) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4153 | - | 1e-108 | 61.44% (306) | hypothetical protein MUL_4153 |
| M. vanbaalenii PYR-1 | Mvan_5335 | - | 1e-134 | 75.99% (304) | hypothetical protein Mvan_5335 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3608|M.bovis_AF2122/97 ------------------MPTARSDAPLSVTWMGVATLLVDDGSSALMTD
Rv3577|M.tuberculosis_H37Rv ------------------MPTARSDAPLSVTWMGVATLLVDDGSSALMTD
MMAR_5077|M.marinum_M MRLRFGRPDLKRYSHRFGVPAAKPDAPLSAMWIGVSTVLLDDGSSALMTD
MUL_4153|M.ulcerans_Agy99 MRLRFGRPDLKRYSHRFGVPAAKPDAPLSAMWIGVSTVLLDDGSSALMTD
MAV_0579|M.avium_104 MRLRLGRPDITRYADRFDVPAAGPGG-LSVTWLGVASLLIDDGSSALLTD
Mflv_1449|M.gilvum_PYR-GCK MRCKPGRPDLDAYATYFDLPAATPASPVTVSWAGVTTLLINDGTSALMTD
Mvan_5335|M.vanbaalenii_PYR-1 MRLKLGRPDLQAYSAYFDLPPAPPGAPVTVTWAGVTTLLVDDGESAVMTD
MSMEG_6071|M.smegmatis_MC2_155 MRLKPGRPDLADHARFFDAPTATPDSPVTVTWAGVTTLLVDDGTSALLTD
TH_1750|M.thermoresistible__bu MRIRPGRPSLTPYAHRFDVPAAAATSRLTVTWAGVTTLLIDDGKSALLTD
*.* . . ::. * **:::*::** **::**
Mb3608|M.bovis_AF2122/97 GYFSRPGLARVAAG-KVSPSAERVDGCLARANVSRLTAVIPVHTHIDHAM
Rv3577|M.tuberculosis_H37Rv GYFSRPGLARVAAG-KVSPSAERVDGCLARANVSRLTAVIPVHTHIDHAM
MMAR_5077|M.marinum_M GYFSRPSLAQVAAG-KLAPSAARVDGCLAQAKASELAAVIPVHTHIDHAM
MUL_4153|M.ulcerans_Agy99 GYFSRPSLAQVAAG-KLAPSAARVDGCLAQAKASELAAVIPVHTHIDHAM
MAV_0579|M.avium_104 GYFSRPGLARVVAG-KVSPSPPRVDGCLARAKVSRLEAVIPVHTHFDHAL
Mflv_1449|M.gilvum_PYR-GCK GFFSRPGLLRVATRRPLASSDSRIDAALQRLGVTRLDGVTPVHTHFDHAM
Mvan_5335|M.vanbaalenii_PYR-1 GFFSRPSHLSVAAR-PLRSSRSRIDDALSRLGVRRLDAVTPVHTHYDHAM
MSMEG_6071|M.smegmatis_MC2_155 GFFSRPGLLTVGLR-PLAPNVPRIDGCLARLGVDRLAAVLPVHTHFDHAM
TH_1750|M.thermoresistible__bu GFFSRPALATVALR-RLEPDTARIEGSLARLRLHRLAAVIPVHTHYDHVL
*:****. * : .. *:: .* : .* .* ***** **.:
Mb3608|M.bovis_AF2122/97 DSALVADRTGAQLVGGESAANVGRGYGLPEESLVVAVPGEPIQLGAFDVT
Rv3577|M.tuberculosis_H37Rv DSALVADRTGAQLVGGESAANVGRGYGLPEESLVVAVPGEPIQLGAFDVT
MMAR_5077|M.marinum_M DSALVADRTGALLVGGESAANIGRGYGLAEDRLVVAVPGQSMELGGYDVT
MUL_4153|M.ulcerans_Agy99 DSALVADRTGALLVGGESAANIGRGYGLADDRLVVAVPGQSMELGAYDVT
MAV_0579|M.avium_104 DSALVADRTGALLVGGESAANVGRGYGLPEDRIVVAADGEPIRLGAFDVT
Mflv_1449|M.gilvum_PYR-GCK DSAAVAGRTGARLIGGSSAAHIGRGHGL--DRVEVVTSGDATAVGAFDVT
Mvan_5335|M.vanbaalenii_PYR-1 DSAVVADLTGARLIGGRSAAAIGRGHGL--DRVDVVAPAETVSVGSYDIT
MSMEG_6071|M.smegmatis_MC2_155 DSAVVAERTGAALVGGSSAAQIGIGGGLPDDRIVTVTPGEPVAMEAYDVT
TH_1750|M.thermoresistible__bu DSPAVADRTGAVLIGGASAAQVGRGAGLPDDRIVVAADGASVSAGSFDIT
**. ** *** *:** *** :* * ** : : ... . . .:*:*
Mb3608|M.bovis_AF2122/97 LVESHHCPPDRFPGVISAPLTPPVKASAYRCGEAWSTLVHHRPSGRRLLI
Rv3577|M.tuberculosis_H37Rv LVESHHCPPDRFPGVISAPLTPPVKASAYRCGEAWSTLVHHRPSGRRLLI
MMAR_5077|M.marinum_M LIESHHCPPDRFPGVISEPVVPPVKASAYRCGEAWSTHIHHRPSGRRLLI
MUL_4153|M.ulcerans_Agy99 LIESHHCPPDRFPGVISEPVVPPVKASAYRCGEAWSTHIHHRPSGRRLLI
MAV_0579|M.avium_104 LVKSHHCPPDRFPGVIAEPLIPPARASAYRCGESWSTLVRHRPTGRRLLI
Mflv_1449|M.gilvum_PYR-GCK LIESHHCPPDRFPGAITAPVAPRASVSAYRCGEAWSTLVRHRGTGRGLLI
Mvan_5335|M.vanbaalenii_PYR-1 LVEGSHCPPDRFPGTITAPVISPARVSAYRCGEAWSTLVHHRPTGVGLLV
MSMEG_6071|M.smegmatis_MC2_155 LFVSEHCPPDRFPGVITEPVVPPAKASAYKCGEAWSTLVHHRPSDRRVLI
TH_1750|M.thermoresistible__bu LIDTGHCPPDRFPGTIDAPVAPPARVSAYRCGEAWSLLVGHRPSGRRLLI
*. *********.* *: . . .***:***:** : ** :. :*:
Mb3608|M.bovis_AF2122/97 QDSAGFVSGALAGYRADAAYLSVGQLGLQPPSYLLEYWTETVRTVGVRRV
Rv3577|M.tuberculosis_H37Rv QDSAGFVSGALAGYRADAAYLSVGQLGLQPPSYLLEYWTETVRTVGVRRV
MMAR_5077|M.marinum_M QGSAGFVAGALDGYRAEVVYLGVGQLGLQRRSYLIDYWNEVVRAVAARRV
MUL_4153|M.ulcerans_Agy99 QGSAGFVAGALDGYRAEVVYLGVGQLGLQRRSYLIDYWNEVVRAVAARRV
MAV_0579|M.avium_104 CGSAGFVPGALAGQRAGAAYLSVGQLGLRPRSYLDEYWDQTVRTVGARRV
Mflv_1449|M.gilvum_PYR-GCK VGSAGFVPGALAGRHADVVYLGVGQLGLQPHDYLAAYWTETVRTVGARRV
Mvan_5335|M.vanbaalenii_PYR-1 VGSAGFVPDALAGRRADVVYLGVGQLGLQPEGYLVDYWTQTVRTVGARQV
MSMEG_6071|M.smegmatis_MC2_155 VGSAGAVPGALAGQRADVVYLGVGQLGRQPERYLTDYWTQTVRTVGARRV
TH_1750|M.thermoresistible__bu VGSAGFRRGALAGQHAEVAYLGVGQLGLQPERYLVDYWTETVRTVGARRV
.*** .** * :* ..**.***** : ** ** :.**:*..*:*
Mb3608|M.bovis_AF2122/97 ILIHWDDFFRPLSKPLRALPYAADDLDLSIRILDELAAQDGVALQMPTVW
Rv3577|M.tuberculosis_H37Rv ILIHWDDFFRPLSKPLRALPYAADDLDLSIRILDELAAQDGVALQMPTVW
MMAR_5077|M.marinum_M VLVHWDDFFRPLSKPMRAFPYAADDLDMSIRILDELAAQDGIPLQMPTVW
MUL_4153|M.ulcerans_Agy99 VLVHWDDFFRPLSKPMRAFPYAADDLDMSIRILDELAAQDGIPLQMPTVW
MAV_0579|M.avium_104 ILIHWDDFFRPLSKPLRALPYAGDDLDSSIRVLDELAERDGVALHLPSVW
Mflv_1449|M.gilvum_PYR-GCK VLIHWDDFFRPLDKPLRALPFVADNLDVSMRILARLAREDGVDLHLPTLW
Mvan_5335|M.vanbaalenii_PYR-1 VLIHWDDFFRPLDKPLRSLPFAVDDLDVSMRVLSRLAAEDGVGLHLPTLW
MSMEG_6071|M.smegmatis_MC2_155 VLIHWDDFFRPLHRPLRALPYVADDLDATMRVLTTLAAHDGVALHLPTLW
TH_1750|M.thermoresistible__bu VLTHWDDFFRPLHRPVRALPYAADDLDFSMRVLGGLAETDGIPLHLPTVW
:* ********* :*:*::*:. *:** ::*:* ** **: *::*::*
Mb3608|M.bovis_AF2122/97 RREDPWM--------
Rv3577|M.tuberculosis_H37Rv RREDPWM--------
MMAR_5077|M.marinum_M QREDPWV--------
MUL_4153|M.ulcerans_Agy99 QREDHWV--------
MAV_0579|M.avium_104 RREDPWA--------
Mflv_1449|M.gilvum_PYR-GCK ERADPWR--------
Mvan_5335|M.vanbaalenii_PYR-1 QPAAPWS--------
MSMEG_6071|M.smegmatis_MC2_155 QRSDPWIETGSGPWR
TH_1750|M.thermoresistible__bu QRTDPWR--------
. *