For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RYVLPDLAETRTDSPFPGLTTHAYLWTPANILFYSPATDADFDDLAALGGVSDQYLSHRDEAGPMLRQIA ERFGTTLHAPLGDRADIEKHAHVDVPLSGHYVDYNGVEVIPTPGHSPGAVCYLVTGAAGRYLFTGDTLFR SAEGHWYAGYIEGFHHLDDADTIAASLRTLADLTPDFVISSAFQGESAVHRIDADAWRGHIAHAIAGLPN SART*
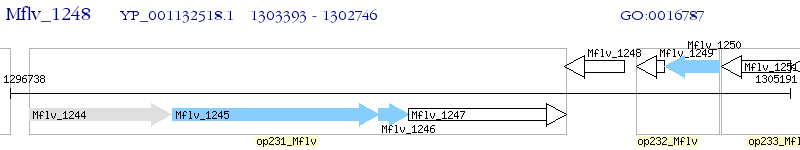
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1248 | - | - | 100% (215) | hypothetical protein Mflv_1248 |
| M. gilvum PYR-GCK | Mflv_5123 | - | 4e-05 | 33.70% (92) | beta-lactamase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0651c | - | 6e-06 | 31.50% (127) | glyoxalase II |
| M. tuberculosis H37Rv | Rv0634c | - | 6e-06 | 31.50% (127) | glyoxalase II |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0965 | gloB | 3e-05 | 35.87% (92) | glyoxalase II, GloB |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3509 | - | 9e-60 | 56.74% (215) | hypothetical protein MSMEG_3509 |
| M. thermoresistible (build 8) | TH_1674 | - | 1e-71 | 62.26% (212) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0715 | gloB | 2e-05 | 35.87% (92) | glyoxalase II, GloB |
| M. vanbaalenii PYR-1 | Mvan_5553 | - | 1e-102 | 80.84% (214) | hypothetical protein Mvan_5553 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0965|M.marinum_M MSDSDRLYFRQLLAGRDFAVNDMIAAQMRNFAYLIGDR---ETGDCVVVD
MUL_0715|M.ulcerans_Agy99 MSDSDRLYFRQLLSGRDFAVNDMIAAQMRNFAYLIGDR---ETGDCVVVD
Mb0651c|M.bovis_AF2122/97 MS-KDRLYFRQLLSGRDFAVGDMFATQMRNFAYLIGDR---TTGDCVVVD
Rv0634c|M.tuberculosis_H37Rv MS-KDRLYFRQLLSGRDFAVGDMFATQMRNFAYLIGDR---TTGDCVVVD
Mflv_1248|M.gilvum_PYR-GCK --------MRYVLPDLAETRTDSPFPGLTTHAYLWTP------ANILFYS
Mvan_5553|M.vanbaalenii_PYR-1 --------MKQVLRDLWETRTDSPFPGLTTHAYLWTP------ANILFYS
TH_1674|M.thermoresistible__bu --------MHQVLPTLWQTRTDSPFPGLTTHAFLWTGG----RRNALFYS
MSMEG_3509|M.smegmatis_MC2_155 ----MTNTMTQIRSDLWQTRTDSPFPGLTTHSYLWKPNGHSSSPGVLFYS
: : : * . : ..::* . :. .
MMAR_0965|M.marinum_M PAYAAG-DLVNALE-ADDMHLSGVLVTHHHPDHVGGSMMGFELKGLAELL
MUL_0715|M.ulcerans_Agy99 PAYAAG-DLVNALE-AGDMHLSGVLVTHHHPDHVGGSMMGFELKGLAELL
Mb0651c|M.bovis_AF2122/97 PAYAAG-DLLDALE-SDDMQLSGVLVTHHHPDHVGGSMMGFQLPGLAELL
Rv0634c|M.tuberculosis_H37Rv PAYAAG-DLLDALE-SDDMQLSGVLVTHHHPDHVGGSMMGFQLPGLAELL
Mflv_1248|M.gilvum_PYR-GCK PATDADFDDLAALGGVSDQYLS-------HRDEAG--------PMLRQIA
Mvan_5553|M.vanbaalenii_PYR-1 PATDAQFNELTELGGVNDQYLS-------HRDEAG--------PMLARIA
TH_1674|M.thermoresistible__bu VATDADFDAIEQLGGIADQYLS-------HRDEAG--------PMLARIA
MSMEG_3509|M.smegmatis_MC2_155 PATDADFAAIDALGGVGHQYLS-------HQDEAG--------PMLARIA
* * : * . ** * *..* * .:
MMAR_0965|M.marinum_M ERVSVPVHVNTHEAVWVSRVTGIPMSDLTTHEHRDKVSVGDIEIELLHTP
MUL_0715|M.ulcerans_Agy99 ERVSVPVHVNTHEAVWVSRVTGIPMSDLTTHEHRDKVSVGDIEIELLHTP
Mb0651c|M.bovis_AF2122/97 ERASVPVHVNTHEALWVSRVTGIPVGDLITHEHGDKVSVGDIDIELLHTP
Rv0634c|M.tuberculosis_H37Rv ERASVPVHVNTHEALWVSRVTGIPVGDLITHEHGDKVSVGDIDIELLHTP
Mflv_1248|M.gilvum_PYR-GCK ERFGTTLHAPLGDRADIEKHAHVDVP-------LSGHYVDYNGVEVIPTP
Mvan_5553|M.vanbaalenii_PYR-1 ERFGSTLHAPAADLADIGKHAHVDVP-------LKGRYVDYNGVEIIPTP
TH_1674|M.thermoresistible__bu RRFGSRLHAPAAESAEISAHATIDVP-------LDRRHVDDNGIEVIPTP
MSMEG_3509|M.smegmatis_MC2_155 EHYGARLHAPAAELANIARHAKVHVP-------LDRRHRDDNGVEVIPTP
.: . :*. : : : : : . . :*:: **
MMAR_0965|M.marinum_M GHTPGSQCFLLDG-----RLVAGDTLFLEGCG---RTDFPGGD----SDE
MUL_0715|M.ulcerans_Agy99 GHTPGSQCFLLDG-----QLVAGDTLFLEGCG---RTDFPGGD----SDE
Mb0651c|M.bovis_AF2122/97 GHTPGSQCFLLDG-----RLVAGDTLFLEGCG---RTDFPGGD----SDE
Rv0634c|M.tuberculosis_H37Rv GHTPGSQCFLLDG-----RLVAGDTLFLEGCG---RTDFPGGD----SDE
Mflv_1248|M.gilvum_PYR-GCK GHSPGAVCYLVTGAAG-RYLFTGDTLFRSAEGHWYAGYIEGFHHLDDADT
Mvan_5553|M.vanbaalenii_PYR-1 GHSPGSVSYLVSGADG-RYLFTGDTLFRNPDGHWWAGYIEGFHQPGDADT
TH_1674|M.thermoresistible__bu GHTPGSVCYLVAGSGGARYLFTGDTLYVDASGAWTAGYLPGMS---DARS
MSMEG_3509|M.smegmatis_MC2_155 GHSPGSTSYLVDGADG-RYLFTGDTVFVDHAGRWSTFVIPGIG---DPAD
**:**: .:*: * *.:***:: . * : * .
MMAR_0965|M.marinum_M IFRSLQQLAALPGDPTVFPGHWYSAEPSASLSEVKRSNFVYRAADLAQWR
MUL_0715|M.ulcerans_Agy99 IFRSLQQLAALPGDPTVFPGHWYSAEPSASLSEVKRSNFVYRAADLAQWR
Mb0651c|M.bovis_AF2122/97 MYRSLRQLAELPGDPTVFPGHWYSAEPSASLSEVKRSNYVYRPASLDQWR
Rv0634c|M.tuberculosis_H37Rv MYRSLRQLAELPGDPTVFPGHWYSAEPSASLSEVKRSNYVYRPASLDQWR
Mflv_1248|M.gilvum_PYR-GCK IAASLRTLADLTPDFVISSAFQGESAVHRIDADAWRGHIAHAIAGLPNSA
Mvan_5553|M.vanbaalenii_PYR-1 IAESLRTLAEVSPDFVISSAFQGNSAVHRVEPDLWRGHIAHAIAGLPTSA
TH_1674|M.thermoresistible__bu LDHSLELLATLTPDVVISSAFAGDSAVHPVNRNRWPEHVEQARAGLAT--
MSMEG_3509|M.smegmatis_MC2_155 MAESLRLLATLEPDVVISSAFGG-AAVSTIDDRPWARCVDEAMGSLAA--
: **. ** : * .: ... : ..*
MMAR_0965|M.marinum_M MLMGG
MUL_0715|M.ulcerans_Agy99 MLMGG
Mb0651c|M.bovis_AF2122/97 MLMGG
Rv0634c|M.tuberculosis_H37Rv MLMGG
Mflv_1248|M.gilvum_PYR-GCK RT---
Mvan_5553|M.vanbaalenii_PYR-1 RR---
TH_1674|M.thermoresistible__bu RR---
MSMEG_3509|M.smegmatis_MC2_155 -----