For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIKDKHQIVIVGGGTAGISVAARLLRNGYSDVAVIEPSSVHYYQPLWTLVGGGQAKAATTARPESSVMPA GATWIKNAAAAVDPDNNTVTCADGAVYEYDVLVVAPGIQLDWNRTEGLADALGKDGVSSNYRFDLAPRTW EFIRDLRSGTAVFAMPSGPIKCAGAPQKIAYLASDYWRSQGVLDNIDVHLVVPTPRLFGIPAIADTLDTI AAGYGITVHTDAEVTSIDAAAHKVGVTPAGGGADTVLAYDVLHAVPRQSAPDWIKSGPLSTGDANGYVEI DKHTMQHVRYPNVFSLGDAGSSPNSKTGAAIRKQAPVVVENIDALLTGRPLTASYNGYGSCPIVTSSHEM LLAEFDYDLNMTPSFPLLDPTKPHRGYWYLKKYGLPFMYWNLMLKGLA
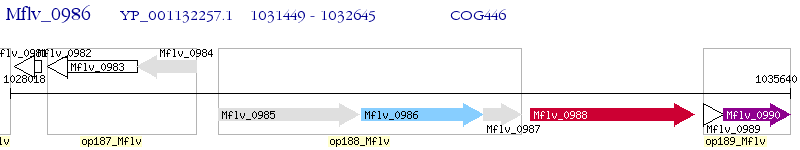
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0986 | - | - | 100% (398) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. gilvum PYR-GCK | Mflv_5068 | - | 2e-05 | 26.58% (301) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0338 | - | 6e-20 | 23.87% (377) | dehydrogenase/reductase |
| M. tuberculosis H37Rv | Rv0331 | - | 6e-20 | 23.87% (377) | dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_02061 | - | 9e-05 | 25.71% (245) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4743c | - | 1e-25 | 26.39% (379) | putative dehydrogenase/reductase |
| M. marinum M | MMAR_1017 | - | 1e-05 | 25.57% (305) | ferredoxin reductase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3457 | - | 6e-06 | 28.33% (120) | DoxD family protein/pyridine nucleotide-disulfide |
| M. thermoresistible (build 8) | TH_2797 | - | 0.0 | 74.62% (394) | PUTATIVE FAD-dependent pyridine nucleotide-disulphide |
| M. ulcerans Agy99 | MUL_0769 | - | 2e-06 | 34.62% (104) | ferredoxin reductase |
| M. vanbaalenii PYR-1 | Mvan_1831 | - | 0.0 | 87.25% (400) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0986|M.gilvum_PYR-GCK ----------------MIKDKHQIVIVGGGTAGISVAARLLR----NGYS
Mvan_1831|M.vanbaalenii_PYR-1 ----------------MTTGKHKILIVGGGTAGISVAARLLR----KGHT
TH_2797|M.thermoresistible__bu ----------VTMMSTRTTARHRILIIGGGTAGITVAARLLR----RGHT
Mb0338|M.bovis_AF2122/97 -------------------MSKTVLILGAGVGGLTTADTLRQLLPPED--
Rv0331|M.tuberculosis_H37Rv -------------------MSKTVLILGAGVGGLTTADTLRQLLPPED--
MAB_4743c|M.abscessus_ATCC_199 --------------------MKKIVILGAGIGGLSVINEIRQSDVPLDGV
MMAR_1017|M.marinum_M MYEHHVTSSEGVTQDPGATAANGIVIIGGGLAAARTAEQLRRAG-YEGQL
MUL_0769|M.ulcerans_Agy99 MYEHHVTSSEGVTQDPGATAANGIVIIGGGLAAARTAEQLRRAG-YEGQL
MLBr_02061|M.leprae_Br4923 -------MNAQPAATAAQDRRHQVVIIGSGFGGLNAAKKLKRAN-----V
MSMEG_3457|M.smegmatis_MC2_155 ---------------------MKVLVIGGGFGGLFCARRLGRLD-----V
::::*.* .. : :
Mflv_0986|M.gilvum_PYR-GCK DVAVIEPSSVHYYQPLWTLVGGGQAKAATTAR--PESSVMPAGATWIKNA
Mvan_1831|M.vanbaalenii_PYR-1 DVAVIEPSEVHYYQPLWTLVGGGQAKASTTAR--AEASVMPKGATWIKNA
TH_2797|M.thermoresistible__bu DVAVIEPSETHYYQAMWTLVGGGQAQAVRTAR--PQAAVMPKGATWIKKS
Mb0338|M.bovis_AF2122/97 RIILVDRSFDGTLGLSLLWVLRGWRRPDDVRV--RPTAASLPGVEMVTAT
Rv0331|M.tuberculosis_H37Rv RIILVDRSFDGTLGLSLLWVLRGWRRPDDVRV--RPTAASLPGVEMVTAT
MAB_4743c|M.abscessus_ATCC_199 DITVVDEDFSHFLGFTLPWVMRGWREVESVPI--RPNAESLSGVTAITGS
MMAR_1017|M.marinum_M TIVSDEVHLPYDRPPLSKEVLRSEVDDVSLKP--REWYDEKDIALRLGSA
MUL_0769|M.ulcerans_Agy99 TIVSDEVHLPYDRPPLSKEVLRSEVDDVSLKP--REWYDEKGITLRLGSA
MLBr_02061|M.leprae_Br4923 DIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNIQVLLGN
MSMEG_3457|M.smegmatis_MC2_155 DVTLVDRAAGHLFQPLLYQCATGTLSIGHISRPLREELARYSNVKPLLGE
: . :
Mflv_0986|M.gilvum_PYR-GCK AAAVDPDNNTVTCA----DGAVYEYDVLVVAPGIQLDWNRTEGLADALGK
Mvan_1831|M.vanbaalenii_PYR-1 AAQVDPDNNRVICV----DGADYGYDVLVVCPGIQLDWNRTEGLADAVGK
TH_2797|M.thermoresistible__bu AATVDPAAGTVGCT----DGSSYGYDVLVMSPGIQLDWDRTDGLVDALGH
Mb0338|M.bovis_AF2122/97 VAHIDIAAQVVHT-----DNSVIGYDALVIALGAALNTDAVPGLSDALDA
Rv0331|M.tuberculosis_H37Rv VAHIDIAAQVVHT-----DNSVIGYDALVIALGAALNTDAVPGLSDALDA
MAB_4743c|M.abscessus_ATCC_199 VTAIDPNAHTVTLS----DSSTLSFDALVIATGARNALDKVRGLAESAAR
MMAR_1017|M.marinum_M ATGLDTAAQTVTLA----DGTVLGYDELVIATGLVP--RRIPAFPELEG-
MUL_0769|M.ulcerans_Agy99 ATGLDTAAQTVTLA----DGTVLGYDELVIATGLVP--RRIPAFPELEG-
MLBr_02061|M.leprae_Br4923 VTHIDLAN-QCVVSELLGHTYETLYDSLIVAAGAGQSYFGNDQFAEFAPG
MSMEG_3457|M.smegmatis_MC2_155 AIRLDPDQRQVTFLRPDETSFVLDYDVLVVAAGMRQSYMGNERFSEWAPG
. :* :* *::. * : :
Mflv_0986|M.gilvum_PYR-GCK DGVSSNYRFDLAPRTWEFIRDLRSGTAVFAMPSGPIKCAGAPQKIAYLAS
Mvan_1831|M.vanbaalenii_PYR-1 DGVSSNYRFDLAPRTWEFIRTLRSGSAVFAMPSGPIKCAGAPQKIAYLAA
TH_2797|M.thermoresistible__bu DGVSSNYRYDLAPRTWEFIRTTRSGTAVFAMPSGPIKCAGAPQKIAYLAC
Mb0338|M.bovis_AF2122/97 DVAGQFYTLDGAAELRAKVEALEHGRIAVAIAGVPFKCPAAPFEAAFLIA
Rv0331|M.tuberculosis_H37Rv DVAGQFYTLDGAAELRAKVEALEHGRIAVAIAGVPFKCPAAPFEAAFLIA
MAB_4743c|M.abscessus_ATCC_199 GDAVHYYSVAGAGDARRALADFSAGKLVFLVTSQPFRCPVAPYEGALLAS
MMAR_1017|M.marinum_M ------------------IRVLRSFDECMALRSHASAAKRAVVIGAGFIG
MUL_0769|M.ulcerans_Agy99 ------------------IRVLRSFDECMALRSHASAAKRAVVIGAGFIG
MLBr_02061|M.leprae_Br4923 MKSIDDALELRGRILSAFEQAERSNDPERREKLLTFTVVGAGPTGVEMAG
MSMEG_3457|M.smegmatis_MC2_155 MKTLDDALSIRQRVFTAFEIAETLPPGPERDAWLTFAVAGGGPTGVELAG
. . . :
Mflv_0986|M.gilvum_PYR-GCK DYWRSQGVLDNIDVHLVVPT--PRLFGI----------PAIADTLDTIAA
Mvan_1831|M.vanbaalenii_PYR-1 DYWRSQGVLQDIDVHLVVPT--PRLFGI----------PAIADNLDKVAA
TH_2797|M.thermoresistible__bu DHWRREGVLSHIDVHLVLPT--PRAFGI----------PEIADSLEEIIA
Mb0338|M.bovis_AF2122/97 AQLGDRYATGTVQIDTFTPDPLPMPVAG----------PEVGEALVSMLK
Rv0331|M.tuberculosis_H37Rv AQLGDRYATGTVQIDTFTPDPLPMPVAG----------PEVGEALVSMLK
MAB_4743c|M.abscessus_ATCC_199 DLLNEKNVRSAVEISMYTPEVHPMPSAG----------PYAGPQLVQLLK
MMAR_1017|M.marinum_M CEVAASLRGLGVEVVLVEPQPTPLAAVLG---------EQIGELVARLHR
MUL_0769|M.ulcerans_Agy99 CEVAASLRGLGVEVVLVEPQPTPLAAVLG---------EQIGELVARLHR
MLBr_02061|M.leprae_Br4923 QIAELADHTLKGAFRRIDSTKARVVLLDAAPAVLPPMGEKLGQRAAARLQ
MSMEG_3457|M.smegmatis_MC2_155 QIREMATRTLANEFHSIEPEEARVLLFDGGDRVLKSFAPDLAERAAESLS
. . .
Mflv_0986|M.gilvum_PYR-GCK GYGITVHTDAEVTSIDAAAHKVGVTPAG--GGADTVLAYDVLHA-VPRQS
Mvan_1831|M.vanbaalenii_PYR-1 GYGITVHTSAEVTAIDAAAHKVALTPVGEAGGSETVLPYDVLHA-VPRQS
TH_2797|M.thermoresistible__bu GYGIHLHTNTEVTAVDGGARKVALTSLGD-GGTDTTLPYDLLHI-APRQS
Mb0338|M.bovis_AF2122/97 DHGVGFHPRKALARVDEAARTMHF-------GDGTSEPFDLLAV-VPPHV
Rv0331|M.tuberculosis_H37Rv DHGVGFHPRKALARVDEAARTMHF-------GDGTSEPFDLLAV-VPPHV
MAB_4743c|M.abscessus_ATCC_199 DNGITFHGERTTECIDPERKVVVF-------GDGTTASFDLLVF-VPPHE
MMAR_1017|M.marinum_M SEGVDVRTGVGVAEVRGDGHVDTVVL-----ADGTQLDADLVVVGIGSHP
MUL_0769|M.ulcerans_Agy99 SEGVDVRTGVGVAEVRGDGHVDTVVL-----ADGTQLDADLVVVGIGSHP
MLBr_02061|M.leprae_Br4923 KMGVEIQLSAMVTDVDRNGITVQDSDG---TVRRIESACKVWSAGVSASR
MSMEG_3457|M.smegmatis_MC2_155 ALGVELRMGVHVTDVRRDGVTVTPKNGG--PAEEYATRTVLWTAGVEAVP
*: .: : :
Mflv_0986|M.gilvum_PYR-GCK APDWIKSGPLSTGDANGYVEIDKHTMQHVRYPNVFSLGDAGSS-------
Mvan_1831|M.vanbaalenii_PYR-1 APDWIKASPLSTGDANGYVEIDKHTLQHVRHRNVFSLGDAGSS-------
TH_2797|M.thermoresistible__bu APDWIKASPLSTGDAGGYVDVDKHTLQHVRYPNVFSLGDAAST-------
Mb0338|M.bovis_AF2122/97 PSAAARSAGLSE---SGWIPVDPRTLSTS-ADNVWAIGDATVLTLPN---
Rv0331|M.tuberculosis_H37Rv PSAAARSAGLSE---SGWIPVDPRTLSTS-ADNVWAIGDATVLTLPN---
MAB_4743c|M.abscessus_ATCC_199 PSVTIDGQ--------GWIAVDAETMQTQ-YDGIWAIGDTAAVTSPS---
MMAR_1017|M.marinum_M ATGWLDGSGIAVDN-----GVVCDAAGRTSAPNVWALGDVASWRDQMGHQ
MUL_0769|M.ulcerans_Agy99 ATGWLDGSGIAVDN-----GVVCDAAGRTSAPNVWAFGDVASWRDQMGHQ
MLBr_02061|M.leprae_Br4923 LGRDLAEQSLVELDWAGRVKVLPDLSIPG-YRNVFVVGDMAAI-------
MSMEG_3457|M.smegmatis_MC2_155 FARHVAEVLGAKTDRAGRIEVEPDLTVPG-HPEVFVVGDLVGR-------
: :: .**
Mflv_0986|M.gilvum_PYR-GCK ----------PNSKTGAAIRKQAPVVVENIDALLTGR----PLTASYNGY
Mvan_1831|M.vanbaalenii_PYR-1 ----------PNSKTGAAIRKQAPVVVENIDAYLAGR----PLTASYNGY
TH_2797|M.thermoresistible__bu ----------PNSKTGAAIRKQAPVVVDNVDAFLAGR----PITAHYDGY
Mb0338|M.bovis_AF2122/97 --------GKPLPKAAVFAEAQAAVVAHGVARHLGYD----VAERHFTGT
Rv0331|M.tuberculosis_H37Rv --------GKPLPKAAVFAEAQAAVVAHGVARHLGYD----VAERHFTGT
MAB_4743c|M.abscessus_ATCC_199 --------GKPLPKAAIFAKNGAKAAAQNVLHYLGRS----DRKAVLSGL
MMAR_1017|M.marinum_M VRVEHWSNVADQARVVVPAMLGREVSSNVVVPYFWSD----QYDVKIQCL
MUL_0769|M.ulcerans_Agy99 VRVEHWSNVADQARVVVPAMLGREVSSNVVVPYFWSD----QYDVKIQCL
MLBr_02061|M.leprae_Br4923 ---------DGVPGVAQGAIQGAKYVANNIKAELGEANPAEREPFQYFDK
MSMEG_3457|M.smegmatis_MC2_155 ---------DKLPGVAENAMQGGLHVARCVRRDLAGK---PRRPYRYHDL
. . : :
Mflv_0986|M.gilvum_PYR-GCK GSCPIVTSSHEMLLAE---FDYDLNMTPSFPLLDPTKPHRGYWYLKKYGL
Mvan_1831|M.vanbaalenii_PYR-1 GSCPLVTSSHDMLLAE---FDYDLNLEPSFPLLDPTKPHRAYWYLKKYGL
TH_2797|M.thermoresistible__bu ASCPIVTSARSVLLAE---FDYSFAMKPTFPLLDPARPHRAYWYLKKYGL
Mb0338|M.bovis_AF2122/97 GACYVETGDHQAAKGD---GDFFAPSAPSVTLYPPSREFHEEKVAQELAW
Rv0331|M.tuberculosis_H37Rv GACYVETGDHQAAKGD---GDFFAPSAPSVTLYPPSREFHEEKVAQELAW
MAB_4743c|M.abscessus_ATCC_199 GYCYLDTGQHRAAQGK---GDFFTLPHPAITLTEPSVGQHHDKRDEEAQW
MMAR_1017|M.marinum_M GEPHSTDVVHLVEDDGRKFLAYYERDGVLVGVVGGGVPGKVMKVRAKIAA
MUL_0769|M.ulcerans_Agy99 GEPHSTDVVHLVEDDGRKLLAYYQRDGVLVGVVGGGVPGKVMKVRAKIAA
MLBr_02061|M.leprae_Br4923 GSMATVSRFSAVAKIGPLEFSGFIAWLMWLVLHLMYLIGFKTKITTLLSW
MSMEG_3457|M.smegmatis_MC2_155 GSAAYIRRGDALLQVHRVRVSGFAGWLAWGFIHIAFLTGMRNRVSTVITW
. :
Mflv_0986|M.gilvum_PYR-GCK PFMYWNLMLKGLA-------------------------------------
Mvan_1831|M.vanbaalenii_PYR-1 PFMYWNLMLKGLA-------------------------------------
TH_2797|M.thermoresistible__bu PFMYWNLMLKGLV-------------------------------------
Mb0338|M.bovis_AF2122/97 LTRWKT--------------------------------------------
Rv0331|M.tuberculosis_H37Rv LTRWKT--------------------------------------------
MAB_4743c|M.abscessus_ATCC_199 RDMWESRSGA----------------------------------------
MMAR_1017|M.marinum_M AVPISEMLDESQG-------------------------------------
MUL_0769|M.ulcerans_Agy99 AVPISEMLDESQGQTSARIPARGAGPGGPLVRWPTLTKSGRWRAEKATAS
MLBr_02061|M.leprae_Br4923 TVTFLSTRR-------GQLTITEQQAFARTRLEQLAELTAESQSTAAANR
MSMEG_3457|M.smegmatis_MC2_155 MATIARASRYHRAFMLGDPTKIEQERYTWSRWDQESPP-PETWPPPAVEQ
Mflv_0986|M.gilvum_PYR-GCK ---------------------------
Mvan_1831|M.vanbaalenii_PYR-1 ---------------------------
TH_2797|M.thermoresistible__bu ---------------------------
Mb0338|M.bovis_AF2122/97 ---------------------------
Rv0331|M.tuberculosis_H37Rv ---------------------------
MAB_4743c|M.abscessus_ATCC_199 ---------------------------
MMAR_1017|M.marinum_M ---------------------------
MUL_0769|M.ulcerans_Agy99 SPVARVDGIGLEPGPRRTASWPDVPGT
MLBr_02061|M.leprae_Br4923 ATMRAS---------------------
MSMEG_3457|M.smegmatis_MC2_155 GP-------------------------