For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARWSRGTTLQNLICHSDAGSQFTSIRYGERLAEIGGVPSIGTVGDSFDNALAETVNGYYKAELIYGPARS GPWKTVEDVELATLSWVYWHNTSRLHSYLGDLPPAEFEAAFYDASRSGQPLVEIQ*
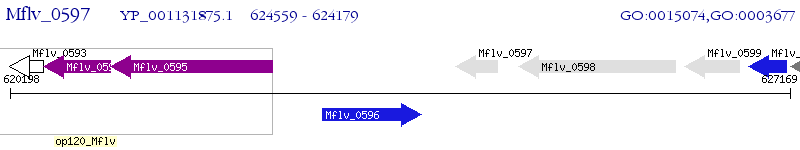
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0597 | - | - | 100% (126) | integrase, catalytic region |
| M. gilvum PYR-GCK | Mflv_0665 | - | 2e-28 | 52.68% (112) | integrase catalytic subunit |
| M. gilvum PYR-GCK | Mflv_3221 | - | 4e-23 | 47.79% (113) | integrase catalytic subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2838c | - | 3e-32 | 60.58% (104) | transposase |
| M. tuberculosis H37Rv | Rv3475 | - | 3e-32 | 60.58% (104) | transposase IS6110 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2086c | - | 2e-54 | 83.76% (117) | putative transposase-like protein IS1601-D |
| M. marinum M | MMAR_2548 | - | 3e-26 | 51.89% (106) | transposase, ISMyma03_aa2 |
| M. avium 104 | MAV_5037 | - | 2e-67 | 92.86% (126) | transposase |
| M. smegmatis MC2 155 | MSMEG_1721 | - | 3e-22 | 46.90% (113) | IS1137, transposase orfB |
| M. thermoresistible (build 8) | TH_3317 | - | 1e-31 | 54.55% (121) | PUTATIVE Integrase, catalytic region |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5973 | - | 1e-22 | 87.04% (54) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2838c|M.bovis_AF2122/97 --------------------------------------------------
Rv3475|M.tuberculosis_H37Rv AEALAAGQRRIAKGERDFKDRVGFLRGRARPASTLITRFIADHQGHREGP
Mflv_0597|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5037|M.avium_104 --------------------------------------------------
Mvan_5973|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3317|M.thermoresistible__bu -----------------------------------MIAFIDAHR------
MMAR_2548|M.marinum_M --------------------------------------------------
MSMEG_1721|M.smegmatis_MC2_155 ------------------------------------MEFISAHQHMRVGV
Mb2838c|M.bovis_AF2122/97 -------------------MPIAPSTYYDHINRE-PSRRELRDGELKEHI
Rv3475|M.tuberculosis_H37Rv DGLRWGVESICTQLT-ELGVPIAPSTYYDHINRE-PSRRELRDGELKEHI
Mflv_0597|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5037|M.avium_104 ---------------------MALSTYYDTKARV-PSARALRDAVLGPAL
Mvan_5973|M.vanbaalenii_PYR-1 ---------------------MAPSTYYDVKSRC-PSARARRDAELGPAL
TH_3317|M.thermoresistible__bu --DQFGVELICRVLRAAIAGFLTSRGYRAAKARP-PSERAMRDELLIAEL
MMAR_2548|M.marinum_M -------------MLTEHGIKISPSTYYEWMKRT-PSKQQLRDEAVTASI
MSMEG_1721|M.smegmatis_MC2_155 DGLKWGVESMC-AVLSEYGVTIAPSTYYAHRARQGPSKADWADAQVIDAI
Mb2838c|M.bovis_AF2122/97 SRVHAAN--YGVYGARKVWLTLNREGIEVARCTVERLMTKLGLSGTTRGK
Rv3475|M.tuberculosis_H37Rv SRVHAAN--YGVYGARKVWLTLNREGIEVARCTVERLMTKLGLSGTTRGK
Mflv_0597|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5037|M.avium_104 CQLWKDN--YCVYGARKLWKTARREGHDVGRDQVARLMRAAGIEGVRRGK
Mvan_5973|M.vanbaalenii_PYR-1 RVIWEDN--YRVYGAVKLWKAARRGGHDVGRDQVARLMRSLGIEGVRRGK
TH_3317|M.thermoresistible__bu RTVHEQN--FSVYGVKKMHHAMKRRGWRVGREQTRRLMRKAGLRGVQRGK
MMAR_2548|M.marinum_M KEFRERNPLNKTLGSRKVWIKLRGDGMDVARCTVERLMQANRWQGARYGG
MSMEG_1721|M.smegmatis_MC2_155 WQLRQSQSLYRVLGARKTWIVLRTNGIDVSRCVVERVMREMGWRGACKRR
Mb2838c|M.bovis_AF2122/97 ARRTTIADPATARPADLVQRRFGPPAPNRLWVADLTYVSTWAGFAYVAFV
Rv3475|M.tuberculosis_H37Rv ARRTTIADPATARPADLVQRRFGPPAPNRLWVADLTYVSTWAGFAYVAFV
Mflv_0597|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2086c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5037|M.avium_104 RVRTTKADPAAARHPDLVHRNFAAAAPNQLWVTDLTFVPTWAGVAYVCFI
Mvan_5973|M.vanbaalenii_PYR-1 RVTTTRADPAAPRHPDLVKRQFTATAPNQLWVTDLTFVPTWVGVAYVCFI
TH_3317|M.thermoresistible__bu PVFTTISDPAHCRPADLVNRQFSAEAPDRLWVADITFVRTWQGFCYTAFV
MMAR_2548|M.marinum_M APKTTIQDEKAQRATDLVDRVFAAPAPNRLWVADFTYCPTWAGMVYVAFV
MSMEG_1721|M.smegmatis_MC2_155 RVRTTVADPAATRAPDRVRRNFVAGAPDRLWVADFTYCRTRAGWAYTAFV
Mb2838c|M.bovis_AF2122/97 TDAYARRILGWRVASTMATS-MVLDAIEQAIWTRQQEGVLDLKDVIHHTD
Rv3475|M.tuberculosis_H37Rv TDAYARRILGWRVASTMATS-MVLDAIEQAIWTRQQEGVLDLKDVIHHTD
Mflv_0597|M.gilvum_PYR-GCK ----------------------------MARWSRGT----TLQNLICHSD
MAB_2086c|M.abscessus_ATCC_199 -----------------------------------------MPGLTCHSD
MAV_5037|M.avium_104 IDAHSRMIVGWRVASHMRTS-MVLDALEMARWSRGT----TLQDLICHSD
Mvan_5973|M.vanbaalenii_PYR-1 VDAFSRLIVGWRAASNMRTT-MVLDALEMARWSRGN----TLSGLRCHSD
TH_3317|M.thermoresistible__bu TDACTKAIKGWAVAASMRTEDLPLEAFNHAVWHSDS----DLSELIHHSD
MMAR_2548|M.marinum_M IDAFARRVIGWRAATSMKTA-LVIDALEHAFFTRAQEGFTDLSGLVAHSD
MSMEG_1721|M.smegmatis_MC2_155 TDVYARKIVGWKVATEMTQK-LVTDAIDHAIDTRKRSGAASLDSLIHHSD
: : *:*
Mb2838c|M.bovis_AF2122/97 RGSQYTSIRFSERLAEAGIQPSVGAVGSSYDNALAETINGLYKTELIKP-
Rv3475|M.tuberculosis_H37Rv RGSQYTSIRFSERLAEAGIQPSVGAVGSSYDNALAETINGLYKTELIKP-
Mflv_0597|M.gilvum_PYR-GCK AGSQFTSIRYGERLAEIGGVPSIGTVGDSFDNALAETVNGYYKAELIYGP
MAB_2086c|M.abscessus_ATCC_199 AGSQFTSIRYGERLAEIGAVPSIGTIGDSYDNAL-ETVNGYYKAELIYGP
MAV_5037|M.avium_104 AGSQFTSIRYGERLAEIGAVPSIGTVGDSFDNALAETVNGYYKAELIYGP
Mvan_5973|M.vanbaalenii_PYR-1 AGSQFTSIRYGERLAELGAVPSIGSVGDSFDNALAERAC---LIDCVSG-
TH_3317|M.thermoresistible__bu RGSQYLSLTYTDRLAELGIAPSVGSRGDSYDNALAEAVNAAYKTELINR-
MMAR_2548|M.marinum_M AGSQYTSIAFTTRLIDAGIDASVGSVADAYDNALAETTVGSFKNELIRR-
MSMEG_1721|M.smegmatis_MC2_155 AGSQYTAVAFTERLAAEGILPSVGSVGDSFDNALAESVNSSYKTELIDR-
***: :: : ** * .*:*: ..::**** * : :
Mb2838c|M.bovis_AF2122/97 --GKPWRSIEDVELATARWVDWFNHRRLYQYCGDVPPVELEAAYY---AQ
Rv3475|M.tuberculosis_H37Rv --GKPWRSIEDVELATARWVDWFNHRRLYQYCGDVPPVELEAAYY---AQ
Mflv_0597|M.gilvum_PYR-GCK ARSGPWKTVEDVELATLSWVYWHNTSRLHSYLGDLPPAEFEAAFY-DASR
MAB_2086c|M.abscessus_ATCC_199 VRTRPWKTVEDVELATLSWVYWHNTSRLHSYLNDIPPTEFEAAFY-DAHG
MAV_5037|M.avium_104 ARTGPWKTVEDVELATLSWVYWHNTSRLHSYLGDIPPAEFEAAFY-DAYR
Mvan_5973|M.vanbaalenii_PYR-1 -----WA-------------------------------------------
TH_3317|M.thermoresistible__bu --GKPWRSVDDVELATAGWVAWYNQERLHEALGYVPPAEYEAALTGTSHP
MMAR_2548|M.marinum_M --QGPWRDVDHVEIGTLNWVDWFNNERPHEYLADLTPTQAEALHY---RH
MSMEG_1721|M.smegmatis_MC2_155 --QPLYPGATELALGTAEWVAFYNRQRPNGYCQDLTPDRAEALYY---HR
:
Mb2838c|M.bovis_AF2122/97 RQRPAAG----
Rv3475|M.tuberculosis_H37Rv RQRPAAG----
Mflv_0597|M.gilvum_PYR-GCK SGQPLVEIQ--
MAB_2086c|M.abscessus_ATCC_199 TDQPLAGIQ--
MAV_5037|M.avium_104 TDQPLIGIQ--
Mvan_5973|M.vanbaalenii_PYR-1 -----------
TH_3317|M.thermoresistible__bu ASQPTPALATN
MMAR_2548|M.marinum_M KSSLAAAG---
MSMEG_1721|M.smegmatis_MC2_155 QRHPHAEEALR