For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PLTLADVIALPVVQRGRPEVLSSRRWDDTIRWVHVSDLADLSTLLQGGELVLTTGAAMARSPRTYLRGLA EAGAVGVVVEVGAVVPEVPPGVGGIAEQLDLALVVLHRTVRFVEVTEAVHRMIVAEQYDVVEFDRRVHET FTDLSMKRASLSGIVDAAAKLLGEHVVLEDLSHQALVASTGIPADLLTDWARRSRRSPGGGAGAEPWSAT DVGPRFEQWGRLVVPYAPADATRAATVLERAAAALAMHRMVEQNRTGLQHQAQSGLIDDVLRGRVVDDRD AAARAQALGLRSNAQYLPVVVRVARPAEIDDPVAGHRRNVAVLDAVAHTVNACGHTGLFSLRGDGEVGAV LSLKDSRTRSVDQALGALARRIHTEIRRVDGSDRVVVTLGPSARNVTDAIAGLAEAIDTAEVALSLRGDV RDFYRTSDIRLRGLISLLRDDPRIRRFAETELRALLAHDAGAEPSNISVLREYLRLAGNKSALAERLHMS RPALYKRLRGIEDALGVDLDDGESMTSLHVAVMVLDSLTGSG*
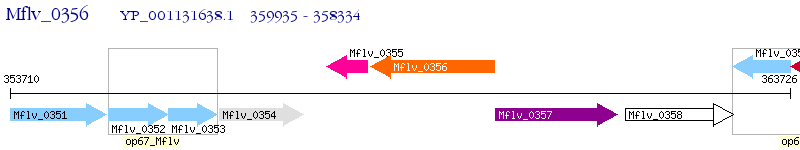
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0356 | - | - | 100% (533) | purine catabolism PurC domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0801 | - | 8e-10 | 27.22% (169) | transcriptional regulator, CdaR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4347 | - | 1e-127 | 46.72% (533) | putative regulatory protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2447 | - | 1e-165 | 58.00% (531) | regulatory protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1825 | - | 1e-166 | 60.43% (513) | purine catabolism PurC domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0356|M.gilvum_PYR-GCK MPLTLADVIALPVVQRGRPEVLSSRRWDDTIRWVHVSDLADLSTLLQGGE
Mvan_1825|M.vanbaalenii_PYR-1 --------------------MLSSGRWDEPIRWVHVGDVPDMSALLLGGE
MSMEG_2447|M.smegmatis_MC2_155 MIPTVRDVIGLPVVQAGDPEVVSAENLDCPVRWVHVSDMPDLAGLLHGGE
MAB_4347|M.abscessus_ATCC_1997 --MTVAEILALPVVLAGDPQVVGGGSLDRPVRWVHVSDVADLSGLLQGGE
::.. * .:*****.*:.*:: ** ***
Mflv_0356|M.gilvum_PYR-GCK LVLTTGAAMAR--SPRTYLRGLAEAGAVGVVVEVGAVVPEVPPGVGGIAE
Mvan_1825|M.vanbaalenii_PYR-1 LVLTTGTALNT--APEVYLRGLADAGALGVVVELGTAMHEVPQSVTDLAR
MSMEG_2447|M.smegmatis_MC2_155 LVLTTGAGLAE--APDDYLERMTRAGAVGLVVELGTRVSRLPRGVSDVAR
MAB_4347|M.abscessus_ATCC_1997 LVLTTGGPLADPDACTRYLGGLFAAGAAGVVVELGTSLQAIPSGVAREAD
****** : : ** : *** *:***:*: : :* .* *
Mflv_0356|M.gilvum_PYR-GCK QLDLALVVLHRTVRFVEVTEAVHRMIVAEQYDVVEFDRRVHETFTDLSMK
Mvan_1825|M.vanbaalenii_PYR-1 SLGLALVALRRQIKFVDVTEAVHRQIVAAQFDEVAFDRKVHETFTELSVR
MSMEG_2447|M.smegmatis_MC2_155 SLGLPLVLLHRQTRFVEITEAVHRLIVADQYEELAFAHRTHETFTDLSMR
MAB_4347|M.abscessus_ATCC_1997 KLQLPVIALHRKIRFVDVTEQVHRSIVAEQYDEVAFARHVHEVFTELTMR
.* *.:: *:* :**::** *** *** *:: : * ::.**.**:*:::
Mflv_0356|M.gilvum_PYR-GCK RASLSGIVDAAAKLLGEHVVLEDLSHQALVASTG--IPADLLTDWARRSR
Mvan_1825|M.vanbaalenii_PYR-1 RASVSSIVDAAARILDQPVVLEDLAHQALAVSARGEPAAELLKNWERRSR
MSMEG_2447|M.smegmatis_MC2_155 RASLADIVRAAADMINESVVLEDLSHQVLAISPRGEAATAVLAEWQRRSR
MAB_4347|M.abscessus_ATCC_1997 RATLGEIVDACGQIAESAIVLEDLNRQVVAFTTHGDSPTGVIHDWERRSR
**::. ** *.. : . :***** :*.:. :. .: :: :* ****
Mflv_0356|M.gilvum_PYR-GCK RS----PGGGAGAEPWSATDVGPRFEQWGRLVVPYAPADATRAATVLERA
Mvan_1825|M.vanbaalenii_PYR-1 RN----PTEGGGAEPWVITSVGLRGEEWGRLLIPSPPAEPGRARMVLERA
MSMEG_2447|M.smegmatis_MC2_155 ----------AMAEPWITTSVGPRTEEWGRLIIPHTPAAPARAKTVLERA
MAB_4347|M.abscessus_ATCC_1997 LTPVLPETGETGPESWLTTPVGPGRQEWGRLIAPSG-GSSLRGRMALERA
.*.* * ** ::****: * . . *. .****
Mflv_0356|M.gilvum_PYR-GCK AAALAMHRMVEQNRTGLQHQAQSGLIDDVLRGRVVDDRDAAARAQALGLR
Mvan_1825|M.vanbaalenii_PYR-1 ATALALHRMIEQNRTGLHQQAQSGLIDDVLQGRIADEREIAARAHALGLR
MSMEG_2447|M.smegmatis_MC2_155 AQALALHRMAERGRSGLEHQAQSGLIEDVLGGRIADDREAAARAEALGLR
MAB_4347|M.abscessus_ATCC_1997 AQALALHRMIEQHQTTLEQQAQSGLVEELRRGRIKDEAEASARARALGVK
* ***:*** *: :: *.:******:::: **: *: : :***.***::
Mflv_0356|M.gilvum_PYR-GCK SNAQYLPVVVRVARPAEIDDPVAGHRRNVAVLDAVAHTVNACGHTGLFSL
Mvan_1825|M.vanbaalenii_PYR-1 KVPRYLPMVVRVDGLAGGVDPVAAQRLNVNLLETVVHTVNASGHTGLFSV
MSMEG_2447|M.smegmatis_MC2_155 AGGPYLPVTVRTEAPAERLDPVGVQRRNIRILDAVTHSVRSQGHTAICSI
MAB_4347|M.abscessus_ATCC_1997 PALSYIPLTVRVRE-EPCADQVLLQRRQIRLLDAVRHAVRTARLSALTAN
*:*:.**. * * :* :: :*::* *:*.: :.: :
Mflv_0356|M.gilvum_PYR-GCK RGDGEVGAVLSLKDSRTRSVDQALGALARRIHTEIRRVDGSDRVVVTLGP
Mvan_1825|M.vanbaalenii_PYR-1 RGPGEIGGLVALNPGR-ASADKALSEIGQQLRRDIGRSTGGKRSVCAVAQ
MSMEG_2447|M.smegmatis_MC2_155 RRDGEIGFVLALTPRRNLTADAALARLAEGLRESVTRSGVTTKVVVAVAN
MAB_4347|M.abscessus_ATCC_1997 PRPGQIDVLLSQTHAS--SAEEILTNVSSVIRASLQRLDDISRCVVGVGP
*::. ::: . :.: * :. :: .: * : * :.
Mflv_0356|M.gilvum_PYR-GCK SARNVTDAIAGLAEAIDTAEVALSLR----GDVRDFYRTSDIRLRGLISL
Mvan_1825|M.vanbaalenii_PYR-1 PGVEIADSIRGLAEAAHIAEVALAMGD---AEARDFHRAADVRLRGLISL
MSMEG_2447|M.smegmatis_MC2_155 NATAFADAVHGLREAAHIAEVALPMAGTARLSGRAFVRASDVRLRGLITL
MAB_4347|M.abscessus_ATCC_1997 ASTRLIDAAVSLDDSAHVAEVALSLPD----DAKAFHRASDTRLRGLLAL
. . *: .* :: . *****.: . : * *::* *****::*
Mflv_0356|M.gilvum_PYR-GCK LRDDPRIRRFAETELRALLAHDAGAEP--SNISVLREYLRLAGNKSALAE
Mvan_1825|M.vanbaalenii_PYR-1 LRDDPRVQQFAETELKAMLAEDPAAHHGLTDLQVLRAYLQLGGNKVALAK
MSMEG_2447|M.smegmatis_MC2_155 MSDDPRVQTFAETELRSLLIHDADHGE--DDMAVLRGYLELAGNKSALAK
MAB_4347|M.abscessus_ATCC_1997 IRSDPRVQAFAEVELRGVLENRARHGD--DLFQLLREFLNTGGNKTELAK
: .***:: ***.**:.:* . . : :** :*. .*** **:
Mflv_0356|M.gilvum_PYR-GCK RLHMSRPALYKRLRGIEDALGVDLDDGESMTSLHVAVMVLDSLTG-----
Mvan_1825|M.vanbaalenii_PYR-1 RLHVSRPALYKRLAAISDKLGVPLDDAESMTSLHVAMMVQDARRRRESPA
MSMEG_2447|M.smegmatis_MC2_155 RLHMSRPALYSRLAAIQRRLGVDLDDGESMTSLHVALLILDTHRSANPEP
MAB_4347|M.abscessus_ATCC_1997 RLHLSRPTLYTKLAIVQRLLGVDLDDAESRTSLHTAMLVLDHVNP-----
***:***:**.:* :. *** ***.** ****.*::: *
Mflv_0356|M.gilvum_PYR-GCK -SG
Mvan_1825|M.vanbaalenii_PYR-1 PSG
MSMEG_2447|M.smegmatis_MC2_155 AR-
MAB_4347|M.abscessus_ATCC_1997 ---