For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSEYPDDIDPDTEPEDGPGTQPAGFADLADDSMATLRPMATQAVFRPHFDDDDDDSDALVHSGDTEPQDA ATTITRHLSPTRRLGGGLVEIPRVPARDPLAALMTDPVVAESKRFCWNCGRPVGRSTKDGRALSEGWCPH CGSAYSFLPQLNPGDIVADQYEIKGCIAHGGLGWVYLAFDKNVNDRPVVLKGLVHSGDAEAQAIAMAERQ FLAEVTHPGIVKIYNFVEHEDKHGNPVGYIVMEYVGGTSLKQATLLKHSSRIRLPAAEAIGFMLEILPAL GYLHSLGLVYNDLKPENIMVTEDQLKIIDLGAVSRINSFGYLYGTPGYQAPEIVRTGPTVATDIYTVGRT LAALTMRLPTRKGRYLDGLPEDNPVLEEYDSFGRLLRRAIDPDPRRRFQTAEELSSQLMGVLREVVASDT GVPRPGLSTMFSPSRSTFGVDLLVAHTDVYLDGQVHSEKLTAQEIVKALQVPLVDPTDVGATVLSATLLS QPVQTLDSLRAVRHGALDSDGVDLSQSVELPLMEVRALLDLGDVAKATRKLDDLAERVGWRWRLVWFRAV SELLTADYDSATKHFTQVLDTLPGELAPKLALAATAELAGASDERTFYKTVWDTDHGVISAGFGLARALS AEGDREGAVRTLDEVPATSRHFTTARLTSAVTLLSGRSTSEITEEHIRSAARRVEALPDTEPRVLQIRAL VLGTAMDWLADNTASTNHILGFPFTEHGLQLGVEASLRSLARVAPTQAHRYALVDLANSVRPMSTF
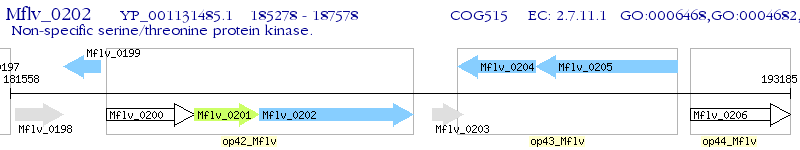
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0202 | - | - | 100% (766) | protein kinase |
| M. gilvum PYR-GCK | Mflv_2972 | - | 2e-22 | 30.03% (303) | protein kinase |
| M. gilvum PYR-GCK | Mflv_1389 | - | 1e-19 | 28.98% (283) | protein kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0418c | pknG | 0.0 | 80.80% (750) | serine/threonine-protein kinase PKNG (protein kinase G) |
| M. tuberculosis H37Rv | Rv0410c | pknG | 0.0 | 80.80% (750) | serine/threonine-protein kinase PKNG (protein kinase G) |
| M. leprae Br4923 | MLBr_00304 | pknG | 0.0 | 78.50% (735) | putative serine-threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_4224 | - | 0.0 | 71.20% (764) | serine/threonine-protein kinase |
| M. marinum M | MMAR_0713 | pknG | 0.0 | 79.63% (756) | serine/threonine-protein kinase PknG |
| M. avium 104 | MAV_4751 | - | 0.0 | 79.24% (766) | serine/threonine protein kinase PknG |
| M. smegmatis MC2 155 | MSMEG_0786 | - | 0.0 | 83.66% (765) | serine/threonine protein kinase |
| M. thermoresistible (build 8) | TH_2307 | pknG | 0.0 | 83.96% (748) | SERINE/THREONINE-PROTEIN KINASE PKNG (PROTEIN KINASE G) |
| M. ulcerans Agy99 | MUL_2810 | pknG | 0.0 | 80.82% (730) | serine/threonine-protein kinase PknG |
| M. vanbaalenii PYR-1 | Mvan_0703 | - | 0.0 | 90.37% (758) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0202|M.gilvum_PYR-GCK ----------------------------------------MSEYPDDIDP
Mvan_0703|M.vanbaalenii_PYR-1 ----------------------------------------MTE-PEDLD-
MSMEG_0786|M.smegmatis_MC2_155 --------------------------------------MTSPENPDLPDA
TH_2307|M.thermoresistible__bu MPETEVNQNHDDARTGTPPAEPTVGSEPTVETRSAETTVGSEQKSTGPAP
MLBr_00304|M.leprae_Br4923 ---------------------------------------MAES-MKREHM
MAV_4751|M.avium_104 ---------------------------------------MAEPDNKSEQP
Mb0418c|M.bovis_AF2122/97 -----------------------------------------------MAK
Rv0410c|M.tuberculosis_H37Rv -----------------------------------------------MAK
MMAR_0713|M.marinum_M ------------------------------MAEAVKKGLQDTSGTENPRP
MUL_2810|M.ulcerans_Agy99 --------------------------------------------------
MAB_4224|M.abscessus_ATCC_1997 ------------------------------------------MSEDPEEP
Mflv_0202|M.gilvum_PYR-GCK DTEPE-DGPGTQPAGFADLADDSMATLRPMATQAVFRPHFDDDDD-DSDA
Mvan_0703|M.vanbaalenii_PYR-1 ------GGQGTQPAGYSDLADDTMSTMRPMATQAVFRPNFFDDEE-DSDA
MSMEG_0786|M.smegmatis_MC2_155 DDAYVDSGPGTQPASLEDLDMDSASTMRPMATQAVYRPEFDDTD----GT
TH_2307|M.thermoresistible__bu ETTSAGPTVGTQPASFDDLGLDTMSTIRPMATQAVFRPHLGDHP----DT
MLBr_00304|M.leprae_Br4923 DHDTEDVGQAAQRADP----PSGTTEGRLQSTQAIFRPNFDDDDD-LLHI
MAV_4751|M.avium_104 EPGTEQVGPGTQPAEVGDDAQGGAATGRLQATQALFRPDFDDDDDDFPHI
Mb0418c|M.bovis_AF2122/97 ASETERSGPGTQPADAQ---TATSATVRPLSTQAVFRPDFGDEDN--FPH
Rv0410c|M.tuberculosis_H37Rv ASETERSGPGTQPADAQ---TATSATVRPLSTQAVFRPDFGDEDN--FPH
MMAR_0713|M.marinum_M AEDEQEAGPGTQPADVQ---TGAAAWSRPIGTQALFRPNFDDDDE--DDI
MUL_2810|M.ulcerans_Agy99 ---------------MQ---TGAAAWSRPIGTQALFRPNFDDDDE--DDI
MAB_4224|M.abscessus_ATCC_1997 DDTSADEGPGTQPASLADLDVDSASTMRPISTRAVFRPPDSTQPA-----
: * .*:*::**
Mflv_0202|M.gilvum_PYR-GCK LVHSGDTEPQDAATTITRHLSPTRRLGGGLVEIPRVPARDPLAALMTDPV
Mvan_0703|M.vanbaalenii_PYR-1 L-HSGDTEPQLTTTTVTRHMSPTRRLGGGLVEIPRVPARDPLAALMTDPV
MSMEG_0786|M.smegmatis_MC2_155 SRGTVVTEAYDQVTMATRALSPMRRLGGGLVEIPRVPERDPLTALMTNPV
TH_2307|M.thermoresistible__bu E-GGDDTEPHDMAVTVARQMSPTRRLGGGLVEIPRVPTKDPLTALMTNPV
MLBr_00304|M.leprae_Br4923 SVPSVDTEPQDRITPATRVLPPIRQLGGGLVEIRRVRDIDPLEALMTNPV
MAV_4751|M.avium_104 SLGALDTDSADRMTVATRALPPVRQLGGGLVEIPRGRDIDPREALMTNPV
Mb0418c|M.bovis_AF2122/97 PTLGPDTEPQDRMATTSRVRPPVRRLGGGLVEIPRAPDIDPLEALMTNPV
Rv0410c|M.tuberculosis_H37Rv PTLGPDTEPQDRMATTSRVRPPVRRLGGGLVEIPRAPDIDPLEALMTNPV
MMAR_0713|M.marinum_M TLGTLDMQPQGPLAAATRARLPVRRLGGGLVEIPRVPDTDPLKALMTDPV
MUL_2810|M.ulcerans_Agy99 TLGTLDMQPQGPLAAATRARLPVRRLGGGLVEIPRVPDTDPLKALMTDPV
MAB_4224|M.abscessus_ATCC_1997 --TQPTTQPEVRATTTRVLLSPTRRLGGGLVEIPRVPEIDPLEALMTNPV
:. . * *:******** * ** ****:**
Mflv_0202|M.gilvum_PYR-GCK VAESKRFCWNCGRPVGRSTKD------GRALSEGWCPHCGSAYSFLPQLN
Mvan_0703|M.vanbaalenii_PYR-1 VAESKRFCWNCGRPVGRSTKD------GRALSEGWCPHCGSAYSFLPQLN
MSMEG_0786|M.smegmatis_MC2_155 VAESKRFCWNCGKPVGRSTPD------GRALSEGWCPHCGSPYSFLPQLS
TH_2307|M.thermoresistible__bu VAESKRFCWNCNRPVGRSKGA------TPAAIEGWCPHCGSPYSFLPQLN
MLBr_00304|M.leprae_Br4923 VPESKRFCWNCGRPVGRSELEGQEADGAQGAKEGWCPYCGSPYSFLPQLS
MAV_4751|M.avium_104 VPESKRFCWNCGKPVGRSTKK------SKGTSEGWCPHCGSAYSFLPQLN
Mb0418c|M.bovis_AF2122/97 VPESKRFCWNCGRPVGRSDSE------TKGASEGWCPYCGSPYSFLPQLN
Rv0410c|M.tuberculosis_H37Rv VPESKRFCWNCGRPVGRSDSE------TKGASEGWCPYCGSPYSFLPQLN
MMAR_0713|M.marinum_M VPEAKRFCWNCGRPVGRSDSE------TKGASEGWCPYCGSPYSFLPQLS
MUL_2810|M.ulcerans_Agy99 VPEAKRFCWNCGRPVGRSDSE------TKGASEGWCPYCGSPYSFLPQLS
MAB_4224|M.abscessus_ATCC_1997 VEESKRFCWNCGKPVGRSTTD------SPAQSEGICPTCGSSFSFLPQLN
* *:*******.:***** . ** ** ***.:******.
Mflv_0202|M.gilvum_PYR-GCK PGDIVADQYEIKGCIAHGGLGWVYLAFDKNVNDRPVVLKGLVHSGDAEAQ
Mvan_0703|M.vanbaalenii_PYR-1 VGDIVADQYEIKGCIAHGGLGWVYLAFDKNVNDRPVVLKGLVHSGDAEAQ
MSMEG_0786|M.smegmatis_MC2_155 PGDIVADQYEIKGCIAHGGLGWVYLAFDKNVNDRPVVLKGLVHSGDAEAQ
TH_2307|M.thermoresistible__bu PGDMVADQYEIKGCIAHGGLGWVYLAFDHNVNERPVVLKGLVHSGDAEAQ
MLBr_00304|M.leprae_Br4923 PGDIVAGQYEVKGCIAHGGLGWVYLAFDHNVNDRPVVLKGLVHSGDAEAQ
MAV_4751|M.avium_104 PGDIVANQYEVKGCIAHGGLGWVYLAVDHNVNDRPVVLKGLVHSGDAEAQ
Mb0418c|M.bovis_AF2122/97 PGDIVAGQYEVKGCIAHGGLGWIYLALDRNVNGRPVVLKGLVHSGDAEAQ
Rv0410c|M.tuberculosis_H37Rv PGDIVAGQYEVKGCIAHGGLGWIYLALDRNVNGRPVVLKGLVHSGDAEAQ
MMAR_0713|M.marinum_M PGDTIAGQYEVKGCIAHGGMGWIYLALDRNVNDRPVVLKGLVHSGDAEAQ
MUL_2810|M.ulcerans_Agy99 PGDTIAGQYEVKGCIAHGGMGWIYLALDRNVNDRPVVLKGLVHSGDAEAQ
MAB_4224|M.abscessus_ATCC_1997 PGDVIAGQYAIKGCIAHGGLGWIYLAVDRNVNDRPVVLKGLVHSGDAEAQ
** :*.** :********:**:***.*:*** *****************
Mflv_0202|M.gilvum_PYR-GCK AIAMAERQFLAEVTHPGIVKIYNFVEHEDKHGNPVGYIVMEYVGGTSLKQ
Mvan_0703|M.vanbaalenii_PYR-1 AIAMAERQFLAQVTHPGIVKIYNFVEHEDKHGNPVGYIVMEYVGGTSLKQ
MSMEG_0786|M.smegmatis_MC2_155 AIAMAERQFLAEVTHPGIVKIYNFVEHEDKHGNPVGYIVMEYVGGTSLKQ
TH_2307|M.thermoresistible__bu AIAMAERQFLAEVTHPGIVKIYNFVEHDDKHGNPVGYIVMEYVGGTSLKE
MLBr_00304|M.leprae_Br4923 ASAVAERQFLAEVVHPQIVQIFNFVEHKDTSGDPVGYIVMEYIGGRSLKR
MAV_4751|M.avium_104 AIAMAERQFLAEVVHPQIVQIFNFVEHVDRHGNPVGYIVMEYVGGQPLRH
Mb0418c|M.bovis_AF2122/97 AMAMAERQFLAEVVHPSIVQIFNFVEHTDRHGDPVGYIVMEYVGGQSLKR
Rv0410c|M.tuberculosis_H37Rv AMAMAERQFLAEVVHPSIVQIFNFVEHTDRHGDPVGYIVMEYVGGQSLKR
MMAR_0713|M.marinum_M AIAMAERQFLAEVVHPSIVQIFNFVEHTDKHGDPVGYIVMEYVGGRSLKR
MUL_2810|M.ulcerans_Agy99 AIAMAERQFLAEVVHPSIVQIFNFVEHTDKHGDPVGYIVMEYVGGRSLKR
MAB_4224|M.abscessus_ATCC_1997 NIAMAERRFLAEVAHPSIVKIFNFVEHPDQHGNPVGYIVMEYVGGTSLKQ
*:***:***:*.** **:*:***** * *:*********:** .*:.
Mflv_0202|M.gilvum_PYR-GCK ATLLKHSSRIRLPAAEAIGFMLEILPALGYLHSLGLVYNDLKPENIMVTE
Mvan_0703|M.vanbaalenii_PYR-1 ATAQKGT---RLPVAEAIGFMLEILPALGHLHSIGLVYNDLKPENVMVTE
MSMEG_0786|M.smegmatis_MC2_155 ARG------AKLPVAEAIGYMLEILPALGYLHSIGLAYNDLKPENIMITE
TH_2307|M.thermoresistible__bu ARD------KKLPVAEAIGYMLEILPALGYLHSIGLAYNDLKPENIMITE
MLBr_00304|M.leprae_Br4923 GSKKGNV--EKLPVAEAIAYLLEILPALSYLHSIGLVYNDLKPENIMLTE
MAV_4751|M.avium_104 G--KG----EKLPVSEAIAYVLEILPALGYLHSIGLVYNDLKPENIMLTE
Mb0418c|M.bovis_AF2122/97 S--KG----QKLPVAEAIAYLLEILPALSYLHSIGLVYNDLKPENIMLTE
Rv0410c|M.tuberculosis_H37Rv S--KG----QKLPVAEAIAYLLEILPALSYLHSIGLVYNDLKPENIMLTE
MMAR_0713|M.marinum_M R--KG----EKLPVSEAIAYLLEILPALAYLHALGLVYNDLKPENIMLTE
MUL_2810|M.ulcerans_Agy99 R--KG----EKLPVSEAIAYLLEILPALAYLHALGLVYNDLKPENIMLTE
MAB_4224|M.abscessus_ATCC_1997 P--KG----NALPVAQAIAYMLEILPALGFLHSVGLTYNDLKPENIMITE
**.::**.::*******..**::**.********:*:**
Mflv_0202|M.gilvum_PYR-GCK DQLKIIDLGAVSRINSFGYLYGTPGYQAPEIVRTGPTVATDIYTVGRTLA
Mvan_0703|M.vanbaalenii_PYR-1 DQLKLIDLGAVSRINSFGYLYGTPGYQAPEIVRTGPTVATDMYTVGRTLA
MSMEG_0786|M.smegmatis_MC2_155 EQLKLIDLGAVSRLNSYGYLYGTPGYQAPEIVRTGPTVATDIYTVGRTLA
TH_2307|M.thermoresistible__bu DQLKLIDLGAVSTINSFGYLYGTPGYQAPEIVRTGPTVQTDIYTVGRTLA
MLBr_00304|M.leprae_Br4923 EQLKLIDLGAVSRINSFGCIYGTPGYQAPEIVRTGPTVATDIYTVGRTLA
MAV_4751|M.avium_104 EQLELIDLGAVSRINSFGYLYGTPGFQAPEIVRTGPTVATDIYTVGRTLA
Mb0418c|M.bovis_AF2122/97 EQLKLIDLGAVSRINSFGYLYGTPGFQAPEIVRTGPTVATDIYTVGRTLA
Rv0410c|M.tuberculosis_H37Rv EQLKLIDLGAVSRINSFGYLYGTPGFQAPEIVRTGPTVATDIYTVGRTLA
MMAR_0713|M.marinum_M EQLKLIDLGAVSRINSFGYLYGTPGFQAPEIVRTGPTVATDIYTVGRTLA
MUL_2810|M.ulcerans_Agy99 EQLKLIDLGAVSRINSFGYLYVTPGFQAPEIVRTGPTVATDIYTVGRTLA
MAB_4224|M.abscessus_ATCC_1997 EQLKLIDLGAVASINAYGNLYGTPGYQAPEISKTGPTVASDIYTVGRTLA
:**::******: :*::* :* ***:***** :***** :*:********
Mflv_0202|M.gilvum_PYR-GCK ALTMRLPTRKGRYLDGLPEDNPVLEEYDSFGRLLRRAIDPDPRRRFQTAE
Mvan_0703|M.vanbaalenii_PYR-1 ALTLKLRTRKGRYVDGLPEDDPVLATYDSYGRLLRRAIDPDPRRRFQSAE
MSMEG_0786|M.smegmatis_MC2_155 ALTLSLRTRRGRYVDGLPSDDPVLETYDSYHRLLRRAIDPDPRRRFTSAE
TH_2307|M.thermoresistible__bu ALTLKLPTRKGRYVDGLPEDDPVLAEYDSFARLLRRATDPDPRRRFASAE
MLBr_00304|M.leprae_Br4923 ALTLNLRTRNGRYMDGLPEDDPVLTTYDSFARLLHRAINPDPRRRFSSAE
MAV_4751|M.avium_104 ALTLNLPTRNGRYVDGIPDNDPVLGAYDSFRRLLRRATDPDPRRRFSSTE
Mb0418c|M.bovis_AF2122/97 ALTLDLPTRNGRYVDGLPEDDPVLKTYDSYGRLLRRAIDPDPRQRFTTAE
Rv0410c|M.tuberculosis_H37Rv ALTLDLPTRNGRYVDGLPEDDPVLKTYDSYGRLLRRAIDPDPRQRFTTAE
MMAR_0713|M.marinum_M ALTLNLPTRNGRYLDGLPDDDPVLASYDSFGRLLRRAIDPDPRRRFASAE
MUL_2810|M.ulcerans_Agy99 ALTLNLPTRNGRYLDGLPDDDPVLASYDSFGRLLRRAIDPDPRRRFASAE
MAB_4224|M.abscessus_ATCC_1997 VLTLRMPTRKGRYKDGLPSDDPVLAKYDSYHRFLRRAIDPDPSARFGSAE
.**: : **.*** **:*.::*** ***: *:*:** :*** ** ::*
Mflv_0202|M.gilvum_PYR-GCK ELSSQLMGVLREVVASDTGVPRPGLSTMFSPSRSTFGVDLLVAHTDVYLD
Mvan_0703|M.vanbaalenii_PYR-1 EMSSQLMGVLREVVAKDTGVPRPGLSTVFSRSRSTFGVDLLVAHTDVYLD
MSMEG_0786|M.smegmatis_MC2_155 EMSSQLLGVLREVVATDTGVPRPGLSTVFSPSRSTFGVDLLVAHTDVYVD
TH_2307|M.thermoresistible__bu EMSAQLLGVLREVVAKDTGVPRPGLSTVFSPTRSTFGIDLLLAHTDVYRD
MLBr_00304|M.leprae_Br4923 EMSAQLMGVLREVVAQDTGVPRAGLSTIFSPSRSTFGVDLLVAHTDVYLD
MAV_4751|M.avium_104 EMSAQLMGVLREVVAHDTGVPRPGLSTIFSPSRSTFGVDLLVAHTDVYLD
Mb0418c|M.bovis_AF2122/97 EMSAQLTGVLREVVAQDTGVPRPGLSTIFSPSRSTFGVDLLVAHTDVYLD
Rv0410c|M.tuberculosis_H37Rv EMSAQLTGVLREVVAQDTGVPRPGLSTIFSPSRSTFGVDLLVAHTDVYLD
MMAR_0713|M.marinum_M EMSGQLMGVLREVVAQDTGVPRPGLSTVFSPSRSTFGVDLLVAHTDVYQD
MUL_2810|M.ulcerans_Agy99 EMSGQLMGVLREVVAQDTGVPRPGLSTVFSPSRSTFGVDLLVAHTDVYQD
MAB_4224|M.abscessus_ATCC_1997 EMATQLVGVLREVVALDSGTPRPGMSTLFSPSRSTFGVDLLVAHTDVYVD
*:: ** ******** *:*.**.*:**:** :*****:***:****** *
Mflv_0202|M.gilvum_PYR-GCK GQVHSEKLTAQEIVKALQVPLVDPTDVGATVLSATLLSQPVQTLDSLRAV
Mvan_0703|M.vanbaalenii_PYR-1 GQVHSEKLTAQEIVRALQVPLVDPTDVGATILSAIVLSQPVQTLDSLRAA
MSMEG_0786|M.smegmatis_MC2_155 GQVHSEKLTAQEIVRALPVPLVDRTDVGAPMLVASVLSEPVHTLDQLRAA
TH_2307|M.thermoresistible__bu GQVHSEKLTAQEIVKALPVPLVDPADVGAPVLTASVLSEPVQTLDQLRAA
MLBr_00304|M.leprae_Br4923 GRLHSEKLTAKDIVTALQVPLVDPTDVAAPVLQATVLSQPVQTLDSLRAA
MAV_4751|M.avium_104 GQVHSEKLTAREIVTALQVPLVDPADVAAPVLQATVLSQPVQTLDSLRAA
Mb0418c|M.bovis_AF2122/97 GQVHAEKLTANEIVTALSVPLVDPTDVAASVLQATVLSQPVQTLDSLRAA
Rv0410c|M.tuberculosis_H37Rv GQVHAEKLTANEIVTALSVPLVDPTDVAASVLQATVLSQPVQTLDSLRAA
MMAR_0713|M.marinum_M GQVHSEKLTAKEIVTALQVPLVDRTDVAASVLQATVLSQPVQTLDSLRAA
MUL_2810|M.ulcerans_Agy99 GQVHSEKLTAKEIVTALQVPLVDRTDVAASVLQATVLSQPVQTLDSLRAA
MAB_4224|M.abscessus_ATCC_1997 GLAHPPSLTAPDIVTALQVPLLDPTDVGAAILQATVLSQPIQTLESLKAA
* *. .*** :** ** ***:* :**.*.:* * :**:*::**:.*:*.
Mflv_0202|M.gilvum_PYR-GCK RHGALDSDGVDLSQSVELPLMEVRALLDLGDVAKATRKLDDLAERVGWRW
Mvan_0703|M.vanbaalenii_PYR-1 RHGTLDSEGIDLSESVELPLMEVRALLDLGDVAKATRKLDDLAERVGWRW
MSMEG_0786|M.smegmatis_MC2_155 RHGALDTEGIDLNESVELPLMEVRALLDLGDVAKATRKLEDLAARVGWRW
TH_2307|M.thermoresistible__bu RHGALDSDGIDLSESVELPLMEVRALLDLGDVAKATRKLDDLAARVGWRW
MLBr_00304|M.leprae_Br4923 RHGMLDAQGIDLAESVELPLMEVRALLDLGDVVKANRKLDDLADRVSCQW
MAV_4751|M.avium_104 RHGTLDADGVELSESIELPLMEVRALLDLGDVAKATRKLDDLAERVGWQW
Mb0418c|M.bovis_AF2122/97 RHGALDADGVDFSESVELPLMEVRALLDLGDVAKATRKLDDLAERVGWRW
Rv0410c|M.tuberculosis_H37Rv RHGALDADGVDFSESVELPLMEVRALLDLGDVAKATRKLDDLAERVGWRW
MMAR_0713|M.marinum_M RHGALDSDGADLSESVELPLMEVRALLDLGDVAKATRKLDDLSERVGWRW
MUL_2810|M.ulcerans_Agy99 RHGALDSDGADLSESVELPLMEVRALLDLGDVAKATRKLDDLSERVGWRW
MAB_4224|M.abscessus_ATCC_1997 RLGRIDADGVDLTESVELPLMEARALLDLGDVAKANSKLDQLEDRVGTPW
* * :*::* :: :*:******.*********.**. **::* **. *
Mflv_0202|M.gilvum_PYR-GCK RLVWFRAVSELLTADYDSATKHFTQVLDTLPGELAPKLALAATAELAGAS
Mvan_0703|M.vanbaalenii_PYR-1 RLVWFRAVSELLTADYDSATKHFNEVLDTLPGELAPKLALAATAELAGSA
MSMEG_0786|M.smegmatis_MC2_155 RLVWFKAVSEMLSADYDSATKHFTEVLDTLPGELAPKLALAATAELAGTA
TH_2307|M.thermoresistible__bu RLVWFRAVAEMLSADYESAMKHFAEVLDTLPGELAPKLALAATAELAGSP
MLBr_00304|M.leprae_Br4923 RLVWYRAVADLLTGDYASATKHFTEVLNTFPGELAPKLALAATAELAGES
MAV_4751|M.avium_104 RLVWYKAVAELLTGDYDSATTHFTEVLDTFPGELAPKLALAATAELAGDV
Mb0418c|M.bovis_AF2122/97 RLVWYRAVAELLTGDYDSATKHFTEVLDTFPGELAPKLALAATAELAGNT
Rv0410c|M.tuberculosis_H37Rv RLVWYRAVAELLTGDYDSATKHFTEVLDTFPGELAPKLALAATAELAGNT
MMAR_0713|M.marinum_M RLVWYRAVSELLTGAYDAAIKHFTEVLDTFPGELAPKLALAATAELAGYT
MUL_2810|M.ulcerans_Agy99 RLVWYRAVSELLTGAYDAAIKHFTEVLDTFPGELAPKLALAATAELAGYT
MAB_4224|M.abscessus_ATCC_1997 RLTWYRGMAALLNGDYDQATKHFTAVLDFLPGEIAPKMALAATAELAGDE
**.*::.:: :*.. * * .** **: :***:***:**********
Mflv_0202|M.gilvum_PYR-GCK DERTFYKTVWDTDHGVISAGFGLARALSAEGDREGAVRTLDEVPATSRHF
Mvan_0703|M.vanbaalenii_PYR-1 DERSFYNTVWSTDNGVISAGFGLARAQSAAGDRDAAVRTLDEVPATSRHF
MSMEG_0786|M.smegmatis_MC2_155 DELKFYKTVWSTDNGVISAGFGLARAQSVAGERDMAVQTLDEVPPTSRHF
TH_2307|M.thermoresistible__bu EAGKFYNTVWSTDHGIISAAFGLARALAAEGDREGAVRVLDQVPATSRHF
MLBr_00304|M.leprae_Br4923 DEHKFYRTVWHTNDGVVSAAFGLARFQSAEGDRTGAVCTLDEVPPTSRHF
MAV_4751|M.avium_104 DEHRFYETVWKTNDGVISAAFGLARTLSAEGDRAAAVRTLDEVPATSRHF
Mb0418c|M.bovis_AF2122/97 DEHKFYQTVWSTNDGVISAAFGLARARSAEGDRVGAVRTLDEVPPTSRHF
Rv0410c|M.tuberculosis_H37Rv DEHKFYQTVWSTNDGVISAAFGLARARSAEGDRVGAVRTLDEVPPTSRHF
MMAR_0713|M.marinum_M DGTNFYQTVWSTNDGVISAAFGLARTLSAQGERAGAVRTLDEVPPSSRHF
MUL_2810|M.ulcerans_Agy99 DGTNFYQTVWSTNDGVISAAFGLARTLSAQGERAGAVRTLDEVPPSSRHF
MAB_4224|M.abscessus_ATCC_1997 KNLDFYRTVWSTDNSVISAGYGLARTLAARGERAEAVRMLDNVPPTSRHF
. **.*** *:..::**.:**** :. *:* ** **:**.:****
Mflv_0202|M.gilvum_PYR-GCK TTARLTSAVTLLSGRSTSEITEEHIRSAARRVEALPDTEPRVLQIRALVL
Mvan_0703|M.vanbaalenii_PYR-1 TTARLTSAVTLLSGRSSSEITEQHIRDAARRVEALPDSEPRVLQIRALVL
MSMEG_0786|M.smegmatis_MC2_155 TTARLTSAVTLLSGRSTSEITEQHIRDAARRVEALPDSEPRVLQIRALVL
TH_2307|M.thermoresistible__bu PTARLTSAVTLLSGRSTSEITEKHIRDAARRVEALPDSEPRVLQIRALVL
MLBr_00304|M.leprae_Br4923 TTARLTSAVTLLSGRSTNEITEQQIRDAARRVETLPPTEPRVLQIRALVL
MAV_4751|M.avium_104 TTARLTSAVTLLSGRSKSEITEEEIRDAARRVEALPPTEPRVLQIRALVL
Mb0418c|M.bovis_AF2122/97 TTARLTSAVTLLSGRSTSEVTEEQIRDAARRVEALPPTEPRVLQIRALVL
Rv0410c|M.tuberculosis_H37Rv TTARLTSAVTLLSGRSTSEVTEEQIRDAARRVEALPPTEPRVLQIRALVL
MMAR_0713|M.marinum_M TTARLTSAVTLLSGRSHGEITEEQIRDAARRVEALPPTEPRVLQIRALVL
MUL_2810|M.ulcerans_Agy99 TTARLTSAVTLLSGRSHGEITEEQIRDAARRVEALPPTEPRVLQIRALVL
MAB_4224|M.abscessus_ATCC_1997 TTARLTSAVTLLSGRTLSEVTEDDIREAARRVEALPETEPRVLQIRALVL
.**************: .*:**..**.******:** :************
Mflv_0202|M.gilvum_PYR-GCK GTAMDWLADNTASTNHILGFPFTEHGLQLGVEASLRSLARVAPTQAHRYA
Mvan_0703|M.vanbaalenii_PYR-1 GTAMDWLADNTASTNHILGFPFTEHGLALGVEASLRSLARVAPTQAHRYA
MSMEG_0786|M.smegmatis_MC2_155 GTALDWLADNTASSNHILGFPFTEHGLKLGVEASLRALARIAPTQSHRYA
TH_2307|M.thermoresistible__bu GTAMDWLADHNASTNHILGFPFTEHGLQLGVESSLRALARVAPTQAHRYA
MLBr_00304|M.leprae_Br4923 GCAMDWLADNQASANHILGFPFTKHGLRLGVEASLRSLARVAPTQRHRYT
MAV_4751|M.avium_104 GCAMDWLEDNKASTNHILGFPFTEHGLRLGVEAALRNLARVAPTQRHRYA
Mb0418c|M.bovis_AF2122/97 GGALDWLKDNKASTNHILGFPFTSHGLRLGVEASLRSLARVAPTQRHRYT
Rv0410c|M.tuberculosis_H37Rv GGALDWLKDNKASTNHILGFPFTSHGLRLGVEASLRSLARVAPTQRHRYT
MMAR_0713|M.marinum_M GGAMDWLQDNEASTNHILGFPFTDHGLRLGVEASLRSLARVAPTQRHRYT
MUL_2810|M.ulcerans_Agy99 GGAMDWLQDNEASTNHILGFPFTDHGLRLGVEASLRSLARVAPTQRHRYT
MAB_4224|M.abscessus_ATCC_1997 GTALDWIKSNHSTGHHILGVPFTKRGLRQGVERCLRALARRATERAHRYA
* *:**: .: :: :****.***.:** *** .** *** *. : ***:
Mflv_0202|M.gilvum_PYR-GCK LVDLANSVRPMSTF
Mvan_0703|M.vanbaalenii_PYR-1 LVDLANSVRPMSTF
MSMEG_0786|M.smegmatis_MC2_155 LVDLANSVRPMSTF
TH_2307|M.thermoresistible__bu LVDLANSVRPMSTF
MLBr_00304|M.leprae_Br4923 LVDMANKVRPTSTL
MAV_4751|M.avium_104 LVDMANKVRPTSTF
Mb0418c|M.bovis_AF2122/97 LVDMANKVRPTSTF
Rv0410c|M.tuberculosis_H37Rv LVDMANKVRPTSTF
MMAR_0713|M.marinum_M LVDMANKVRPTSTF
MUL_2810|M.ulcerans_Agy99 LVDMANKVRPTSTF
MAB_4224|M.abscessus_ATCC_1997 LVDLANATRPTSLL
***:** .** * :