For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SARVLAMAGAVLAGLLTGCSSGHDAVAQGGTFEFVSPGGKTDIYYDPPSSRGRPGPLSGPDLADPAHTLS LDDPIFAGRVVVINIWGQWCGPCRSEVSQLQQVYDATRGAGASFLGIDVRDNNRQAALDFVNDRHVTFPS IYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRELLATDLQPVVQRVAQQ*
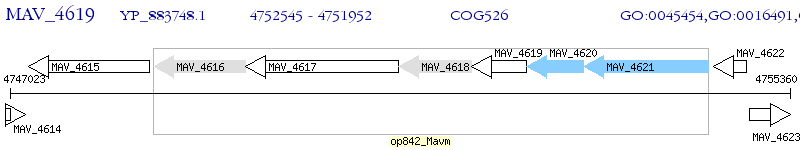
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4619 | - | - | 100% (197) | hypothetical protein MAV_4619 |
| M. avium 104 | MAV_0456 | - | 4e-08 | 32.11% (109) | hypothetical protein MAV_0456 |
| M. avium 104 | MAV_1653 | - | 2e-05 | 29.91% (117) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0539 | - | 1e-92 | 82.74% (197) | thioredoxin protein |
| M. gilvum PYR-GCK | Mflv_0052 | - | 4e-80 | 71.58% (190) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. tuberculosis H37Rv | Rv0526 | - | 1e-92 | 82.74% (197) | thioredoxin protein |
| M. leprae Br4923 | MLBr_02412 | - | 5e-87 | 81.77% (192) | hypothetical protein MLBr_02412 |
| M. abscessus ATCC 19977 | MAB_3976c | - | 3e-74 | 69.59% (194) | thioredoxin |
| M. marinum M | MMAR_0872 | - | 3e-92 | 84.54% (194) | thioredoxin protein (thiol-disulfide interchange protein) |
| M. smegmatis MC2 155 | MSMEG_0971 | - | 3e-83 | 75.13% (189) | hypothetical protein MSMEG_0971 |
| M. thermoresistible (build 8) | TH_2251 | - | 7e-83 | 74.61% (193) | POSSIBLE THIOREDOXIN PROTEIN (THIOL-DISULFIDE INTERCHANGE |
| M. ulcerans Agy99 | MUL_0625 | - | 2e-92 | 83.25% (197) | thioredoxin protein (thiol-disulfide interchange protein) |
| M. vanbaalenii PYR-1 | Mvan_3710 | - | 2e-80 | 72.68% (194) | redoxin domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0539|M.bovis_AF2122/97 --------MQSRATRRSGALTMRRLVIAAAVSALLLTGCS-GRDAVAQGG
Rv0526|M.tuberculosis_H37Rv --------MQSRATRRSGALTMRRLVIAAAVSALLLTGCS-GRDAVAQGG
MAV_4619|M.avium_104 -----------MSAR--------VLAMAGAVLAGLLTGCSSGHDAVAQGG
MLBr_02412|M.leprae_Br4923 --MIAPATMQSEAMRRSGATIIWRLANAGALLATLLAGCS-GRDAVVQGG
MMAR_0872|M.marinum_M -------------MR--------RLVIAAAVLAALLTGCS-GRDAVVQGG
MUL_0625|M.ulcerans_Agy99 -----------MTMR--------RLVIAAAVLAALLTGCS-GRDAVVQGG
MAB_3976c|M.abscessus_ATCC_199 -------------MR--------LLVAALAALALFITACSTGDDAVARGG
Mflv_0052|M.gilvum_PYR-GCK -----------MNRVVAFGRTLMAACAATAVF----AGCATGSDAVAQGG
TH_2251|M.thermoresistible__bu -----------VKRLVVL-----LVGTAMLVL----TGCSTGDDAVAQGG
Mvan_3710|M.vanbaalenii_PYR-1 MGIERWRRRRPPTRWLLVIRRGIAAGLSAVLFGAAVSGCSSGDDAVAQGG
MSMEG_0971|M.smegmatis_MC2_155 -----------MQSWVARVVGVTLAALFAVLTG-ALAGCSTGDDAVAQGG
:.*: * ***.:**
Mb0539|M.bovis_AF2122/97 TFEFVSPGGKTDIFYDPPASRGRPGPLSGPELADPARSVSLDD--FPGQV
Rv0526|M.tuberculosis_H37Rv TFEFVSPGGKTDIFYDPPASRGRPGPLSGPELADPARSVSLDD--FPGQV
MAV_4619|M.avium_104 TFEFVSPGGKTDIYYDPPSSRGRPGPLSGPDLADPAHTLSLDDPIFAGRV
MLBr_02412|M.leprae_Br4923 TFEFVSPDGKTDIFYEPPASRGRPGPLSGLDLADPERTVALDD--FTGNV
MMAR_0872|M.marinum_M TFEFVSPGGKTDIFYDPPASRGHPGPLSGPDLIDPDRTLSLDD--FAGRV
MUL_0625|M.ulcerans_Agy99 TFEFVSPGGKTDIFYDPPASRGHPGPLSGPDLIDPDRTLSLDD--FAGRV
MAB_3976c|M.abscessus_ATCC_199 QFQFVSPGGKTDILYDPPQSRQTPGPISGPSLLDPTKTLSLSD--FRGRV
Mflv_0052|M.gilvum_PYR-GCK TFEFVAPGGQTDIFYDPPQERGTPGPLSGPDLMDTDRTISLDD--FAGKV
TH_2251|M.thermoresistible__bu TFEFVAPGGQTDIFYDPPADRGRPGPISGPDLMNPDETISLDD--FAGQV
Mvan_3710|M.vanbaalenii_PYR-1 TFEFISPGGQVDIFYDPPQSRGRPGPIRGPDLMDPNRTLSLDD--FAEQV
MSMEG_0971|M.smegmatis_MC2_155 TFEFVAPGGKTDIFYDPPQSRGRPGTLAGPLLTDPSKTISLDD--FAGEV
*:*::*.*:.** *:** .* **.: * * :. .:::*.* * .*
Mb0539|M.bovis_AF2122/97 VVVNVWGQWCGPCRAEVSQLQRVYDATRGAGVSFLGIDVRDNNRQAPQDF
Rv0526|M.tuberculosis_H37Rv VVVNVWGQWCGPCRAEVSQLQRVYDATRGAGVSFLGIDVRDNNRQAPQDF
MAV_4619|M.avium_104 VVINIWGQWCGPCRSEVSQLQQVYDATRGAGASFLGIDVRDNNRQAALDF
MLBr_02412|M.leprae_Br4923 VVINVWGQWCGPCRTEVTQLQQVYNATRDSGVSFLGIDVRDNNRQAAQDF
MMAR_0872|M.marinum_M VVVNVWGQWCGPCRAEITQLQQVYDATRAAGVSFLGIDVRDNSRAAAQDF
MUL_0625|M.ulcerans_Agy99 VVVNVWGQWCGPCRAEITQLQQVYDETRAAGVSFLGIDVRDNSRAAAQDF
MAB_3976c|M.abscessus_ATCC_199 VVINVWGQWCGPCRSEFSALEDVYRATHAQGVEFLGINVRDNEITKAQDF
Mflv_0052|M.gilvum_PYR-GCK VVINVWGQWCGPCRTEITELQKVYDATRDRGVAFLGIDVRDNNIDAPRDF
TH_2251|M.thermoresistible__bu VVINIWGQWCGPCRTEILELQQVYDATRHQGVAFLGIDVRDNNIDAAQDF
Mvan_3710|M.vanbaalenii_PYR-1 VVINVWGQWCGPCRTEMPELQRVYDATRNEGVAFLGIDVRDNNRQAAVDF
MSMEG_0971|M.smegmatis_MC2_155 VVVNVWGQWCGPCRAEITQLQEVHNATKDKGVAFLGIDVRDNNRDAAVDF
**:*:*********:*. *: *: *: *. ****:****. . **
Mb0539|M.bovis_AF2122/97 INDRHVTYPSIYDPAMRTLIAFGGKYPTSVIPSTLVLDRQHRVAAVFLRE
Rv0526|M.tuberculosis_H37Rv INDRHVTYPSIYDPAMRTLIAFGGKYPTSVIPSTLVLDRQHRVAAVFLRE
MAV_4619|M.avium_104 VNDRHVTFPSIYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRE
MLBr_02412|M.leprae_Br4923 VNDRQVTFPSIYDPAMRTLIAFGDKYPTTVIPFTLVLDRQHRVAAVFLRE
MMAR_0872|M.marinum_M VRDRHVTFPSIYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRE
MUL_0625|M.ulcerans_Agy99 VRDRHVTFPSIYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRE
MAB_3976c|M.abscessus_ATCC_199 VTDRKVPYPSIYDPSMRTLIAFGKKFPTGAIPATLVLDRQHRVAAVFLRE
Mflv_0052|M.gilvum_PYR-GCK IIDRAITFPSIYDPPMRTMIAFGGRYPTTVIPSTVVLDREHRVAAVFLRE
TH_2251|M.thermoresistible__bu IADRNITFPSIYDPPMRTMIAFGGRYPTTVIPSTVVLDREHRVAAVFLRE
Mvan_3710|M.vanbaalenii_PYR-1 IVDRNVTFPSIYDPPMRTMIALGARYPTTVIPSTIVLDRSHRVAAVFLRA
MSMEG_0971|M.smegmatis_MC2_155 VKDRNVTFPSIYDPPMRTMIAFGGKYPTTVIPSTVVLDRQHRVAAVFLRE
: ** :.:******.***:**:* ::** .** *:****.*********
Mb0539|M.bovis_AF2122/97 LLAADLQPVVERVAEEEPSGRAPVGAQ----
Rv0526|M.tuberculosis_H37Rv LLAADLQPVVERVAEEEPSGRAPVGAQ----
MAV_4619|M.avium_104 LLATDLQPVVQRVAQQ---------------
MLBr_02412|M.leprae_Br4923 LLAGDLQPVVARVAHETVAGQLPRGTS----
MMAR_0872|M.marinum_M LLAEDLQPVVQRLAQEPAAGSTPPGPQ----
MUL_0625|M.ulcerans_Agy99 LLAEDLQPVVQRLAQEPAAGSTPPGPQ----
MAB_3976c|M.abscessus_ATCC_199 LLAQDLRPVVERLAQESAKETAGEVK-----
Mflv_0052|M.gilvum_PYR-GCK LLAQDLQPVVERLAAEQ--------------
TH_2251|M.thermoresistible__bu LLAEDLQPVVERLAAEDPAPDGDAS------
Mvan_3710|M.vanbaalenii_PYR-1 LLAEDLEPVVRRLAAETATG-----------
MSMEG_0971|M.smegmatis_MC2_155 LLAEDLEPLIERLLAEPAEPGEPAVSKEQRP
*** **.*:: *: :