For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSTSLPPLVEPASALSREEVARYSRHLIIPDLGVDGQKRLKNARVLVIGAGGLGAPTLLYLAAAGVGTIG IVDFDVVDESNLQRQVIHGVADVGRSKAQSARDSIVAINPLIRVRLHELRLAPSNAVDLFKQYDLILDGT DNFATRYLVNDAAVLAGKPYVWGSIYRFEGQASVFWEDAPDGLGVNYRDLYPEPPPGMVPSCAEGGVLGI ICASVASVMGTEAIKLITGIGETLLGRLLVYDALEMSYRTITIRKDPSTPKITELVDYEQFCGVVADDAA QAAKGSTITPRELRDWLDSGRKLALIDVRDPVEWDIVHIDGAQLIPKSLINSGEGLAKLPQDRTAVLYCK TGVRSAEALAAVKKAGFSDAVHLQGGIVAWAKQMQPDMVMY
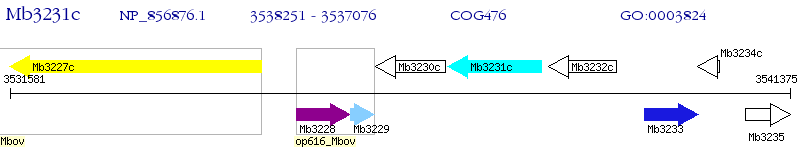
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3231c | moeB1 | - | 100% (391) | molybdopterin biosynthesis-like protein MoeZ |
| M. bovis AF2122 / 97 | Mb3143 | moeB2 | e-119 | 56.01% (391) | molybdenum cofactor biosynthesis protein MoeB |
| M. bovis AF2122 / 97 | Mb2366c | moeW | 3e-12 | 26.02% (246) | hypothetical protein Mb2366c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4670 | - | 0.0 | 81.38% (392) | molybdopterin biosynthesis-like protein MoeZ |
| M. tuberculosis H37Rv | Rv3206c | moeB1 | 0.0 | 99.74% (392) | molybdopterin biosynthesis-like protein MoeZ |
| M. leprae Br4923 | MLBr_00817 | moeZ | 0.0 | 86.19% (391) | molybdopterin biosynthesis-like protein MoeZ |
| M. abscessus ATCC 19977 | MAB_3525c | - | 0.0 | 83.80% (389) | molybdenum cofactor biosynthesis protein MoeB1 |
| M. marinum M | MMAR_1352 | moeB1 | 0.0 | 88.24% (391) | molybdenum cofactor biosynthesis protein MoeB1 |
| M. avium 104 | MAV_4153 | - | 1e-180 | 79.40% (398) | molybdopterin biosynthesis-like protein MoeZ |
| M. smegmatis MC2 155 | MSMEG_1937 | - | 0.0 | 82.91% (392) | molybdopterin biosynthesis-like protein MoeZ |
| M. thermoresistible (build 8) | TH_0450 | moeZ | 0.0 | 80.87% (392) | PROBABLE MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN MOEB1 |
| M. ulcerans Agy99 | MUL_2527 | moeB1 | 0.0 | 87.72% (391) | molybdopterin biosynthesis-like protein MoeZ |
| M. vanbaalenii PYR-1 | Mvan_1797 | - | 0.0 | 81.36% (397) | molybdopterin biosynthesis-like protein MoeZ |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4670|M.gilvum_PYR-GCK ----MSTPLPPLVEPAAELTKEEVARYSRHLIIPDLGLDGQKRLKNARVL
Mvan_1797|M.vanbaalenii_PYR-1 ----MSTPLPPLVEPAAELTREEVARYSRHLIIPDVGLDGQKRLKNARVL
MSMEG_1937|M.smegmatis_MC2_155 MSANLDVSLPPLVEPADELTREEVARYSRHLIIPDLGLDGQKRLKNAKVL
MAB_3525c|M.abscessus_ATCC_199 ----MPT-LPPLVRPAAELTREEVARYSRHLIIPDLGVDGQKRLKNAKVL
TH_0450|M.thermoresistible__bu ----VSSPLPPLVEPAAELTREEVARYSRHLIIPDVGVEGQKRLKNARVL
Mb3231c|M.bovis_AF2122/97 ---MS-TSLPPLVEPASALSREEVARYSRHLIIPDLGVDGQKRLKNARVL
Rv3206c|M.tuberculosis_H37Rv ---MS-TSLPPLVEPASALSREEVARYSRHLIIPDLGVDGQKRLKNARVL
MMAR_1352|M.marinum_M ---MSTTALPPLVAPAATLSREEVARYSRHLIIPDLGVDGQKRLKNARVL
MUL_2527|M.ulcerans_Agy99 ---MSTTALPPLVAPAATLSREEVARYSRHLIIPDLGVDGQKRLKNARVL
MLBr_00817|M.leprae_Br4923 -MSTSSTSLPPLVEPAGQLSRDEVIRYSRHLIIPDIGVDGQMRLKSARVL
MAV_4153|M.avium_104 ----MSTPLPPLVAPADQLTADEMARYSRQLIIPGLGVDGQKRLKNARVL
***** ** *: :*: ****:****.:*::** ***.*:**
Mflv_4670|M.gilvum_PYR-GCK VIGAGGLGSPTLLYLAAAGVGTIGIVEFDVVDESNLQRQIIHGQSDIGRP
Mvan_1797|M.vanbaalenii_PYR-1 VIGAGGLGSPTLLYLAAAGVGTIGIVEFDLVDESNLQRQIIHGQSDIGRS
MSMEG_1937|M.smegmatis_MC2_155 VIGAGGLGSPTLLYLAAAGVGTIGIVEFDVVDESNLQRQIIHGQSDIGRS
MAB_3525c|M.abscessus_ATCC_199 VIGAGGLGSPTLLYLAAAGVGTIGIVEFDVVDESNLQRQIIHGQSDIGRS
TH_0450|M.thermoresistible__bu VIGAGGLGSPALLYLAAAGVGTLGVIDFDVVDESNLQRQVIHGQADVGRP
Mb3231c|M.bovis_AF2122/97 VIGAGGLGAPTLLYLAAAGVGTIGIVDFDVVDESNLQRQVIHGVADVGRS
Rv3206c|M.tuberculosis_H37Rv VIGAGGLGAPTLLYLAAAGVGTIGIVDFDVVDESNLQRQVIHGVADVGRS
MMAR_1352|M.marinum_M VIGAGGLGSPTLLYLAAAGVGTIGIVDFDVVDESNLQRQIIHGVADVGRS
MUL_2527|M.ulcerans_Agy99 VIGAGGLGSPTLLYLAAAGVGTIGIVDFDVVDESNLQRQIIHGVADVGRS
MLBr_00817|M.leprae_Br4923 VIGAGGLGAPVLLYLAAAGVGTIGIIDFDVVDESNLQRQIIHGVADVGRS
MAV_4153|M.avium_104 VIGAGGLGAPTLLYLAAAGVGTIGIVEFDAVEESNLQRQIIHGVADVGRS
********:*.***********:*:::** *:*******:*** :*:**.
Mflv_4670|M.gilvum_PYR-GCK KAQSAKDSVLEVNPLVTVNLHEFRLEPDNAVELFEQYDLILDGTDNFATR
Mvan_1797|M.vanbaalenii_PYR-1 KAQSAKDSIAEINPLVNVVLHEFRLEPDNAVDLFAQYDLILDGTDNFATR
MSMEG_1937|M.smegmatis_MC2_155 KAQSARDSIAEINPLVTVNLHEFRLEPDNAVDLFGQYDLILDGTDNFATR
MAB_3525c|M.abscessus_ATCC_199 KAQSARDSVLEINPLVEVRLHELRLDPENAVELFAQYDLILDGTDNFATR
TH_0450|M.thermoresistible__bu KAESARDTIRAINPFVDVRLHQFRLDRSNAVDLFSEYDLILDGTDNFATR
Mb3231c|M.bovis_AF2122/97 KAQSARDSIVAINPLIRVRLHELRLAPSNAVDLFKQYDLILDGTDNFATR
Rv3206c|M.tuberculosis_H37Rv KAQSARDSIVAINPLIRVRLHELRLAPSNAVDLFKQYDLILDGTDNFATR
MMAR_1352|M.marinum_M KAQSARDSIVAINPLIEVRLHEFRLESDNAVDLFGHYDLIIDGTDNFATR
MUL_2527|M.ulcerans_Agy99 KAQSARDSIVAINPLIEVRLHEFRLESDNAVDLFGHYDLIIDGTDNFATR
MLBr_00817|M.leprae_Br4923 KAQSARDSIVAINPLVQVRLHEFRLESSNVVNLFKQYDLIVDGTDNFATR
MAV_4153|M.avium_104 KAASARDSIAAINPLVDVRLHEFRLDASNAVELFGHYDLIVDGTDNFATR
** **:*:: :**:: * **::** .*.*:** .****:*********
Mflv_4670|M.gilvum_PYR-GCK YLVNDAAVLAGKPYVWGSIYRFEGQVSIFWEDAP-----NGLGLNYRDLY
Mvan_1797|M.vanbaalenii_PYR-1 YLVNDAAVLAGKPYVWGSIYRFEGQISVFWEDAPADEAGNERGLNYRDLY
MSMEG_1937|M.smegmatis_MC2_155 YLVNDAAVLAGKPYVWGSIYRFEGQVSVFWEDAP-----DGLGLNYRDLY
MAB_3525c|M.abscessus_ATCC_199 YLVNDAAVLAHKPYVWGSIYRFEGQVSVFWEDAP-----DGLGLNYRDLY
TH_0450|M.thermoresistible__bu YLVNDAAVLAGKPYVWGSIYRFEGQVSVFWEDAP-----NGQGLNYRDLY
Mb3231c|M.bovis_AF2122/97 YLVNDAAVLAGKPYVWGSIYRFEGQASVFWEDAP-----DGLGVNYRDLY
Rv3206c|M.tuberculosis_H37Rv YLVNDAAVLAGKPYVWGSIYRFEGQASVFWEDAP-----DGLGVNYRDLY
MMAR_1352|M.marinum_M YLVNDAAVLAGKPYVWGSIYRFEGQVSVFWEDAP-----GGLGLNYRDLY
MUL_2527|M.ulcerans_Agy99 YLVNDAAVLAGKPYVWGSIYRFEGQVSVFWEDAP-----GGLGLNYRDLY
MLBr_00817|M.leprae_Br4923 YLINDAAVLAKKPYVWGSLYRFEGQVSVFWEDAP-----DGLGLNYRDLY
MAV_4153|M.avium_104 YLINDAAVLAGKPYVWGSIYRFEGQASVCWEDAP-----DGRGLNYRDLY
**:******* *******:****** *: ***** . *:******
Mflv_4670|M.gilvum_PYR-GCK PEPPPPGMVPSCAEGGVLGILCASIASVMGTEAIKLITGIGDPLLGRLMV
Mvan_1797|M.vanbaalenii_PYR-1 PEPPPPGMVPSCAEGGVLGILCASVGSVMGTEAIKLITGIGETLLGRLMV
MSMEG_1937|M.smegmatis_MC2_155 PEPPPPGMVPSCAEGGVLGILCSSIASIMGTEAIKLITGIGEPLLGRLMV
MAB_3525c|M.abscessus_ATCC_199 PEPPPPGMVPSCAEGGVLGILCASIASVMGTEAIKLITGIGDSLLGRLMV
TH_0450|M.thermoresistible__bu PEPPPPGMVPSCAEGGVLGILPATIGAIQGTEAIKLITGIGETLLGQLLI
Mb3231c|M.bovis_AF2122/97 PEPPP-GMVPSCAEGGVLGIICASVASVMGTEAIKLITGIGETLLGRLLV
Rv3206c|M.tuberculosis_H37Rv PEPPPPGMVPSCAEGGVLGIICASVASVMGTEAIKLITGIGETLLGRLLV
MMAR_1352|M.marinum_M PEPPPPGMVPSCAEGGVLGIICASIASVMGTEAVKLITGIGETLLGRLMI
MUL_2527|M.ulcerans_Agy99 PEPPPPGMVPSCAEGGVLGIICASIASVMGTEAVKLITGIGETLLGRLMI
MLBr_00817|M.leprae_Br4923 LEPPPPGMVPSCAEGGVLGIVCASIASVMGTEAIKLITGIGAPLLGRLMI
MAV_4153|M.avium_104 PEPPPPGAVPSCAEGGVLGVVCAAIASVMSTEVIKLITGIGESLLGRLMI
**** * ***********:: :::.:: .**.:******* .***:*::
Mflv_4670|M.gilvum_PYR-GCK YDALDMSYRTIKIRKDP---STPKITELIDYEAFCG---VVSDAAAEAAA
Mvan_1797|M.vanbaalenii_PYR-1 YDALDMTYRTIKIRKDP---STPKITELIDYEAFCG---VVSDAAAEAAA
MSMEG_1937|M.smegmatis_MC2_155 YDALDMTYRTIRIRKDP---STPKITELIDYEEFCG---VVSDEAAAAAA
MAB_3525c|M.abscessus_ATCC_199 YDALDMTYRTIKIRKDP---ATPKITELIDYEAFCG---VISDEASAAVA
TH_0450|M.thermoresistible__bu YDALEMSYRKIRIRKDP---STPAITELIDYEEFCG---VVSDAAAEAAA
Mb3231c|M.bovis_AF2122/97 YDALEMSYRTITIRKDP---STPKITELVDYEQFCG---VVADDAAQAAK
Rv3206c|M.tuberculosis_H37Rv YDALEMSYRTITIRKDP---STPKITELVDYEQFCG---VVADDAAQAAK
MMAR_1352|M.marinum_M YDALEMTYRTIRIRKDP---ATPKITALIDYEQFCG---VVSDDVARAAS
MUL_2527|M.ulcerans_Agy99 YDALEMTYRTIRIRKDP---ATPKITALIDYEQFCG---VVSDDVARAAS
MLBr_00817|M.leprae_Br4923 YNALEMSYRRIRIHKDP---SRPKITELTDYQQFCG---VVSDDAAQVAT
MAV_4153|M.avium_104 YDALEMSYRTIAIRRDPCDASRPAITTLVDYEQLCGAAPAASTDAATGGA
*:**:*:** * *::** : * ** * **: :** . : .:
Mflv_4670|M.gilvum_PYR-GCK DSTITPRELRELLDSGKPLALIDVREPVEWDINRIEGAELIPKGGFESGE
Mvan_1797|M.vanbaalenii_PYR-1 ESTITPRELRELMDSGKPLALIDVREPVEWDINHIEGAELIPKGAFESGE
MSMEG_1937|M.smegmatis_MC2_155 DSTVTPRELKELLDSGKPLALIDVRERVEWDINHIEGAELIPKPTFESGA
MAB_3525c|M.abscessus_ATCC_199 DSTITPRELRELLDTDKKVALIDVREPVEWDIVHIDGAELVPKSTLESGD
TH_0450|M.thermoresistible__bu DSTVTPRELRELLDAGK-VALVDVREPVEWEINRIEGAELIPKSEIESGV
Mb3231c|M.bovis_AF2122/97 GSTITPRELRDWLDSGRKLALIDVRDPVEWDIVHIDGAQLIPKSLINSGE
Rv3206c|M.tuberculosis_H37Rv GSTITPRELRDWLDSGRKLALIDVRDPVEWDIVHIDGAQLIPKSLINSGE
MMAR_1352|M.marinum_M DSTITPQELRAMIDSGKKLALIDVREQVEWDIVHIDGAQLIPKSLISTGE
MUL_2527|M.ulcerans_Agy99 DSTITPQELRAMINSGKKLALIDVREQVEWDIVHIDGAQLIPKSLISTGE
MLBr_00817|M.leprae_Br4923 GSIVTPRELRELLDSGKKLALIDVREPVEWDIVHIDGAQLIPRSLINSGE
MAV_4153|M.avium_104 EAAITPRQLRELLDSGAKLALIDVREPVEFDIVHLDGAQLIPQSSINSGE
: :**::*: :::. :**:***: **::* :::**:*:*: :.:*
Mflv_4670|M.gilvum_PYR-GCK ALSKLDNDRTPVFYCKTGIRSAEVLAIVKKAGFADAMHVQGGIVAWGKQL
Mvan_1797|M.vanbaalenii_PYR-1 ALSKLKVDRTPVFYCKTGIRSAEVLAIVKKAGFSDAMHVQGGIVAWGKQL
MSMEG_1937|M.smegmatis_MC2_155 ALAKLPTDRTPVFYCKTGVRSAEVLAIAKKAGFSDAVHLQGGIVAWAKQL
MAB_3525c|M.abscessus_ATCC_199 GLAKLPQDRQAVLYCKTGIRSAEALVAVKKAGFSDAVHLQGGIAAWAKQV
TH_0450|M.thermoresistible__bu GLAKLPMDRAPVLYCKTGVRSAEALAAVKRAGFADAMHLQGGIVAWAKQL
Mb3231c|M.bovis_AF2122/97 GLAKLPQDRTAVLYCKTGVRSAEALAAVKKAGFSDAVHLQGGIVAWAKQM
Rv3206c|M.tuberculosis_H37Rv GLAKLPQDRTAVLYCKTGVRSAEALAAVKKAGFSDAVHLQGGIVAWAKQM
MMAR_1352|M.marinum_M GLAKLPQDRTTVLYCKTGVRSAEALAALKKAGFADAVHLQGGIVAWANQM
MUL_2527|M.ulcerans_Agy99 GLAKLPQDRTTVLYCKTGVRSAEALAALKKAGFADAAHLQGGIVAWANQM
MLBr_00817|M.leprae_Br4923 GLAKLPQDRMSVLYCKTGVRSAETLATVKKAGFSDAVHLQGGIVAWAKQV
MAV_4153|M.avium_104 GLAKLPADRMPVLYCKTGVRSAQALAVVRQAGFSDAVHLQGGIVAWAQQM
.*:** ** .*:*****:***:.*. ::***:** *:****.**.:*:
Mflv_4670|M.gilvum_PYR-GCK EPDMVMY
Mvan_1797|M.vanbaalenii_PYR-1 EPDMVMY
MSMEG_1937|M.smegmatis_MC2_155 EPDMVMY
MAB_3525c|M.abscessus_ATCC_199 DPDMVMY
TH_0450|M.thermoresistible__bu EPDMVMY
Mb3231c|M.bovis_AF2122/97 QPDMVMY
Rv3206c|M.tuberculosis_H37Rv QPDMVMY
MMAR_1352|M.marinum_M QPDMVMY
MUL_2527|M.ulcerans_Agy99 QPDMVMY
MLBr_00817|M.leprae_Br4923 QPDMVIY
MAV_4153|M.avium_104 QPDMILY
:***::*