For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTALNRAVASARVGTEVIRVRGLTFRYPKAAEPAVRGMEFTVGRGEIFGLLGPSGAGKSTTQKLLIGLLR DHGGQATVWDKEPAEWGPDYYERIGVSFELPNHYQKLTGYENLRFFASLYAGATADPMQLLAAVGLADDA HTLVGKYSKGMQMRLTFARSLINDPELLFLDEPTSGLDPVNARKIKDIIVDLKARGRTIFLTTHDMATAD ELCDRVAFVVDGRIVALDSPTELKIARSRRRVRVEYRGDGGGLETAEFGMDGLADDPAFHSVLRNHHVET IHSREASLDDVFVEVTGRQLT
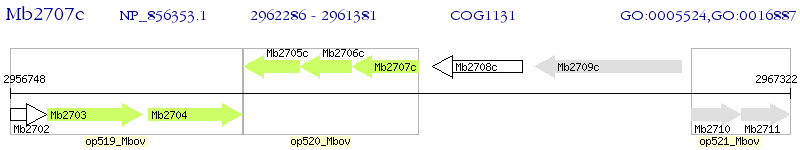
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2707c | - | - | 100% (301) | antibiotic ABC transporter ATP-binding protein |
| M. bovis AF2122 / 97 | Mb1493c | - | 1e-31 | 32.49% (317) | unidentified antibiotic-transport ATP-binding protein ABC |
| M. bovis AF2122 / 97 | Mb1250c | - | 9e-28 | 32.91% (237) | tetronasin-transport ATP-binding protein ABC transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3680 | - | 3e-33 | 32.12% (302) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2688c | - | 1e-172 | 99.67% (301) | antibiotic ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_00590 | - | 5e-34 | 32.35% (306) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1858 | - | 1e-129 | 78.32% (286) | antibiotic ABC transporter ATP-binding protein |
| M. marinum M | MMAR_2026 | - | 1e-134 | 79.79% (292) | antibiotic-transport ATP-binding protein ABC transporter |
| M. avium 104 | MAV_3321 | - | 7e-33 | 31.48% (305) | ABC transporter, ATP-binding subunit |
| M. smegmatis MC2 155 | MSMEG_1502 | - | 1e-135 | 81.12% (286) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_2788 | - | 1e-24 | 29.86% (278) | PUTATIVE ABC transporter ATP-binding protein |
| M. ulcerans Agy99 | MUL_1859 | - | 3e-32 | 32.89% (304) | antibiotic ABC transporter ATP-binding protein |
| M. vanbaalenii PYR-1 | Mvan_0776 | - | 1e-130 | 80.99% (284) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3321|M.avium_104 -----------------MRLRGVSKHYG--STTAVRDLDLEVHAAEVLAL
MUL_1859|M.ulcerans_Agy99 ----------------MVRLRGVGKQYG--SVTAVSNLDLEVRAAEVMAL
MLBr_00590|M.leprae_Br4923 ---MSSACTVPETPEVVIRLRGVCKHYG--SILAVSNLDLDIHSAEVLAL
Mflv_3680|M.gilvum_PYR-GCK MTSVSPGAVPDGSPSAPVVLRGVSKRYG--STTAVAGLDLVVERAEVFAL
Mb2707c|M.bovis_AF2122/97 MTALNRAVASARVGTEVIRVRGLTFRYPKAAEPAVRGMEFTVGRGEIFGL
Rv2688c|M.tuberculosis_H37Rv MTALNRAVASARVGTEVIRVRGLTFRYPKAAEPAVRGMEFTVGRGEIFGL
MSMEG_1502|M.smegmatis_MC2_155 ----------MGSASEVIDVRDLTFTYPRAAEPAVRGMDFAVGNGEIFGF
Mvan_0776|M.vanbaalenii_PYR-1 ----------MNTES-VIDVRDLTYTYPKTAEPAVRGMDFAVGRGEIFGF
MMAR_2026|M.marinum_M --MRDPEPAVSGAVGEVIRVRGLTFAYPKASTPAVRGMDFSVGRGEIFGF
MAB_1858|M.abscessus_ATCC_1997 -------------MTEVIQVRELTFTYPKTPEPAVRGMDFTVGEGEIFGF
TH_2788|M.thermoresistible__bu ------------MMAAMIELTALTKVFG--RTRAVDDLTCTVEPGVVTGF
: : : : ** .: : . : .:
MAV_3321|M.avium_104 LGPNGAGKTTTVEMCEGFVRPDAGTLEVLGLDPIADNARLRPRIGVMLQG
MUL_1859|M.ulcerans_Agy99 LGPNGAGKTTTVEMCEGFVRPDAGSIEVLGLDPIADNARLRPRIGVMLQG
MLBr_00590|M.leprae_Br4923 LGPNGAGKTTTVEMCEGFVRPDAGTIEVLGLDPITDNARLRARIGVMLQG
Mflv_3680|M.gilvum_PYR-GCK LGPNGAGKTTTVEMCEGFIKPDAGSLRILGLDPIVDNAEVRARTGVMLQG
Mb2707c|M.bovis_AF2122/97 LGPSGAGKSTTQKLLIGLLRDHGGQATVWDKEPAEWGPDYYERIGVSFEL
Rv2688c|M.tuberculosis_H37Rv LGPSGAGKSTTQKLLIGLLRDHGGQATVWDKEPAEWGPDYYERIGVSFEL
MSMEG_1502|M.smegmatis_MC2_155 LGPSGAGKSTTQKLLIGLLRGHGGQALVWGRDPVDWGQDYYQRVGVSFEL
Mvan_0776|M.vanbaalenii_PYR-1 LGPSGAGKSTTQKLLIGLLRGHGGQALVWGRDPLDWGPRYYQRIGVSFEL
MMAR_2026|M.marinum_M LGPSGAGKSTTQRILIGLLRGHGGEVAVWGKDPASWGSDYYERIGVSFEL
MAB_1858|M.abscessus_ATCC_1997 LGPSGAGKSTTQKLLIGLLRGHGGRATVWGREPGAWGRNYYQRIGVSFEL
TH_2788|M.thermoresistible__bu LGPNGAGKTTTMRLILGLDRPTSGTATIDGRRYRDLRDPLRT-VGALLDA
***.****:** .: *: : .* : . *. ::
MAV_3321|M.avium_104 GGGYPAARAGEMLNLVAAYAADP-LDPAWLLDTLGLTDAARTTYRRLSGG
MUL_1859|M.ulcerans_Agy99 GGGYPAARAGEMLDLVASYAADP-LDPRWLLDTLGLTDAARTTYRRLSGG
MLBr_00590|M.leprae_Br4923 GGGYPAARAGEMLNLVASYAADP-LDPEWLLETLGLTDVARTTYRRLSGG
Mflv_3680|M.gilvum_PYR-GCK GGAYPAARAGEMLDLVAAYAADP-LDPTWLLDTLGLTDAARTTYRRLSGG
Mb2707c|M.bovis_AF2122/97 PNHYQKLTGYENLRFFASLYAGATADPMQLLAAVGLADDAHTLVGKYSKG
Rv2688c|M.tuberculosis_H37Rv PNHYQKLTGYENLRFFASLYAGATADPMQLLAAVGLADDAHTLVGKYSKG
MSMEG_1502|M.smegmatis_MC2_155 PNHYLRLTGLENLQFFASLYEKQTEDPMELLHAVGLADAAYTRVGEYSKG
Mvan_0776|M.vanbaalenii_PYR-1 PNHYLKLTGAENLQFFASLYDSAGPDPRSLLDDVGLTDAADTRVAKYSKG
MMAR_2026|M.marinum_M PNHYQKLTGLENLQFFASLYSAPTLDPMQLLEAVGLADHADTRAGKYSKG
MAB_1858|M.abscessus_ATCC_1997 PNHYQKLTALENLRFFASLYAGPTRDPMELLDAVGLADDANTRVGAFSKG
TH_2788|M.thermoresistible__bu RAVHPARSARNHLRWLAAANRIPQRRVDEVLELVGLTAVAARPAGTLSLG
: . : * .*: :* :**: * * *
MAV_3321|M.avium_104 QQQRLALACALVGRPELVFLDEPTAGMDAHARLLVWELIDALRRDGVTVV
MUL_1859|M.ulcerans_Agy99 QQQRLALACALVGRPELVFLDEPTAGMDAHARLLVWELIDALRRDGVTVV
MLBr_00590|M.leprae_Br4923 QQQRLALACTLVGRPELVFLDEPTAGMDTHARLLVWELIDTLRRDGVTVV
Mflv_3680|M.gilvum_PYR-GCK QQQRLALACAVVGRPELVFLDEPTAGMDAHARIVVWELIDALRRDGVTVV
Mb2707c|M.bovis_AF2122/97 MQMRLTFARSLINDPELLFLDEPTSGLDPVNARKIKDIIVDLKARGRTIF
Rv2688c|M.tuberculosis_H37Rv MQMRLPFARSLINDPELLFLDEPTSGLDPVNARKIKDIIVDLKARGRTIF
MSMEG_1502|M.smegmatis_MC2_155 MQMRLTFARSLINKPELLFLDEPTSGLDPVNARRVKEIVLDLKSKGRTIF
Mvan_0776|M.vanbaalenii_PYR-1 MQMRLTFARSLVNDPELLFLDEPTSGLDPVNARRIKDIILDLKARGRTVF
MMAR_2026|M.marinum_M MQMRLTFARALLNDPELLFLDEPTSGMDPVNARSVKNMIMDLKARGRTVF
MAB_1858|M.abscessus_ATCC_1997 MQMRLTFVRALINDPELLFLDEPTSGLDPVNARKVKDMVLELKARGRTIF
TH_2788|M.thermoresistible__bu MSQRLGIAGALLGDPPVLLFDEPVNGLDPEGIRWVRTLMRTMAAEGRTVL
. ** :. :::. * ::::***. *:*. : :: : * *:.
MAV_3321|M.avium_104 LTTHQLKEAEELADRLVIIDHGATVAAGTPAELMRSGAKDQLRFSAPPRL
MUL_1859|M.ulcerans_Agy99 LTTHQLKEAEELADRLVIIDHGVTVAEGTPSELMRSGAKDQLRFTAPPRL
MLBr_00590|M.leprae_Br4923 LTTHQLKEAEELADRLVIIDHGRMIAAGTPTELMHTGAKDQLRFSAPPRL
Mflv_3680|M.gilvum_PYR-GCK LTTHQLSEAEELADRILIIDNGAAVATGTPAELMRNGAENQLRFRAPKML
Mb2707c|M.bovis_AF2122/97 LTTHDMATADELCDRVAFVVDGRIVALDSPTELKIARSRRRVRVEYRG--
Rv2688c|M.tuberculosis_H37Rv LTTHDMATADELCDRVAFVVDGRIVALDSPTELKIARSRRRVRVEYRG--
MSMEG_1502|M.smegmatis_MC2_155 LTTHDMSTADELCDRIAFVVDGSIVALDTPAELKIARSRRRVRVQYRV--
Mvan_0776|M.vanbaalenii_PYR-1 LTTHDMATADELCDRVAFVVDGRIVALDAPAELKIARSRRLVRVEYRS--
MMAR_2026|M.marinum_M LTTHDMATADELCDRVAFVVDGSIVVSDRPAELKIARSQRLVTVEYRG--
MAB_1858|M.abscessus_ATCC_1997 ITTHAMSAADELCDRVAFVVDGRIVAMNSPAELKLEYGRRSVRVEYRG--
TH_2788|M.thermoresistible__bu VSSHLLAEMANTADRLLVIGRGRLIASTSVAEFVGSTAAERVRVRSPEMD
:::* : : .**: .: * :. :*: . : .
MAV_3321|M.avium_104 DLSLLSAALPEDYKATELTPGEYLVEGPVDPQVLATVTAWCARIDVLATD
MUL_1859|M.ulcerans_Agy99 DLSLLVSALPEDYRVTEVTPGEYLVEGSVDPQVLAAVTAWCARIDVLATD
MLBr_00590|M.leprae_Br4923 DLSLLAFALPEDYKTTELTPGEYLVAGPIDPQVLATVTAWCAQINVLTTN
Mflv_3680|M.gilvum_PYR-GCK DLSLLVAALPEGYRATEPTAGEYLVEGHIDPQVLATVTAWCARLDVLATD
Mb2707c|M.bovis_AF2122/97 -------------DGGGLETAEFGMDGLADDPAFHSVLR-----NHHVET
Rv2688c|M.tuberculosis_H37Rv -------------DGGGLETAEFGMDGLADDPAFHSVLR-----NHHVET
MSMEG_1502|M.smegmatis_MC2_155 -------------ADGGLRTAEFGMDGLADDPEFHAVLR-----DHHVET
Mvan_0776|M.vanbaalenii_PYR-1 -------------AQGTLQTTEFPMDGLADDPEFRTVLR-----DHHVET
MMAR_2026|M.marinum_M -------------NGGGLSAAEFSLDTLADDPAFHAVLR-----EHQVET
MAB_1858|M.abscessus_ATCC_1997 -------------AAGELAGQDFPLDGLADNDAFHAVLR-----GHHVET
TH_2788|M.thermoresistible__bu TLRG-----LLADAGVEVRLEPDGAALTVRGAAPEVVGELAARNGITLHE
*
MAV_3321|M.avium_104 MRVEQRSLEDVFLDLTGRKLRQ-----
MUL_1859|M.ulcerans_Agy99 MRVEQRSLEDVFLDLTGRKLRS-----
MLBr_00590|M.leprae_Br4923 MRVEQHSLEDVFLDLTGRKLR------
Mflv_3680|M.gilvum_PYR-GCK MRVEQRSLEDVFLDLTGRELRK-----
Mb2707c|M.bovis_AF2122/97 IHSREASLDDVFVEVTGRQLT------
Rv2688c|M.tuberculosis_H37Rv IHSREASLDDVFVEVTGRQLT------
MSMEG_1502|M.smegmatis_MC2_155 IHSREASLDDVFVEVTGRQLA------
Mvan_0776|M.vanbaalenii_PYR-1 IHSREASLDEVFVDVTGRSLA------
MMAR_2026|M.marinum_M IHSREASLEDVFVEVTGRRLS------
MAB_1858|M.abscessus_ATCC_1997 IHSEEASLDDVFVAVTGRRLA------
TH_2788|M.thermoresistible__bu LSVQRASLEDAYLELTDAAVEYRAVPR
: .. **::.:: :*. :