For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRWIGLSTGLVSAMLVAGLVACGSNSPASSPAGPTQGARSIVVFAAASLQSAFTQIGEQFKAGNPGVNVN FAFAGSSELATQLTQGATADVFASADTAQMDSVAKAGLLAGHPTNFATNTMVIVAAAGNPKKIRSFADLT RPGLNVVVCQPSVPCGSATRRIEDATGIHLNPVSEELSVTDVLNKVITGQADAGLVYVSDALSVATKVTC VRFPEAAGVVNVYAIAVLKRTSQPALARQFVAMVTAAAGRRILDQSGFAKP
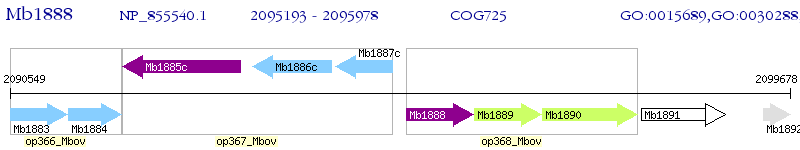
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1888 | modA | - | 100% (261) | molybdate-binding lipoprotein |
| M. bovis AF2122 / 97 | Mb2422c | subI | 5e-09 | 29.34% (167) | sulfate-binding lipoprotein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3859 | - | 9e-72 | 51.72% (261) | molybdenum ABC transporter, periplasmic molybdate-binding |
| M. tuberculosis H37Rv | Rv1857 | modA | 1e-143 | 100.00% (261) | molybdate-binding lipoprotein |
| M. leprae Br4923 | MLBr_00615 | subI | 6e-06 | 20.14% (288) | putative sulphate-binding protein |
| M. abscessus ATCC 19977 | MAB_2433 | - | 3e-67 | 52.33% (258) | molybdenum ABC transporter periplasmic protein |
| M. marinum M | MMAR_2731 | modA | 1e-96 | 70.16% (258) | molybdate-binding lipoprotein ModA |
| M. avium 104 | MAV_2863 | modA | 4e-90 | 65.13% (261) | molybdate ABC transporter, periplasmic molybdate-binding |
| M. smegmatis MC2 155 | MSMEG_2016 | modA | 1e-68 | 57.47% (221) | molybdate ABC transporter, periplasmic molybdate-binding |
| M. thermoresistible (build 8) | TH_1176 | - | 5e-65 | 49.62% (260) | PROBABLE MOLYBDATE-BINDING LIPOPROTEIN MODA |
| M. ulcerans Agy99 | MUL_3022 | modA | 9e-93 | 71.19% (243) | molybdate-binding lipoprotein ModA |
| M. vanbaalenii PYR-1 | Mvan_2538 | - | 6e-69 | 50.97% (257) | molybdenum ABC transporter, periplasmic molybdate-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1888|M.bovis_AF2122/97 -MRWIG-----------LSTGLVSAMLVAGLVA-CGSNSPASSPAGPTQG
Rv1857|M.tuberculosis_H37Rv -MRWIG-----------LSTGLVSAMLVAGLVA-CGSNSPASSPAGPTQG
MMAR_2731|M.marinum_M -MTRVG-----------MLAGMVSALLVW-VVG-CGS------PSRPADG
MUL_3022|M.ulcerans_Agy99 ------------------------------MVG-CGS------PSRPAEG
MAV_2863|M.avium_104 -MRRIG-----------ILTGLLSVVLIAGMTG-CGS------KSQPPPT
Mflv_3859|M.gilvum_PYR-GCK MIRHPA-----------VAVALTVATLMSGCST-AGEDRPAATPPTSDAV
Mvan_2538|M.vanbaalenii_PYR-1 MRSMPG-----------LTSALIAVVLVAGCSS-SRTD-SGPTP------
MSMEG_2016|M.smegmatis_MC2_155 ---MIR-----------VVSVVVGVFLLAGCTP-APAN------------
MAB_2433|M.abscessus_ATCC_1997 MTGSTRNVLTSRAWKVLAATLLAGSLIVSGCGTRTGSGGGSASPSTGAGD
TH_1176|M.thermoresistible__bu MIRVRR------------WLSVCLALMLPAVAG-CGTG--------RTGG
MLBr_00615|M.leprae_Br4923 MFKDIG----RFMRWRHSMVVGVTLASTTDVVAACHGGASDVVGGGTAHA
Mb1888|M.bovis_AF2122/97 ARSIVVFAAASLQSAFTQIGEQFKAGNPG--VNVNFAFAGSSELATQLTQ
Rv1857|M.tuberculosis_H37Rv ARSIVVFAAASLQSAFTQIGEQFKAGNPG--VNVNFAFAGSSELATQLTQ
MMAR_2731|M.marinum_M ARSITIFAAASLKTAFTQIGERFKTANPG--VGVELDFAGSSELATQLTQ
MUL_3022|M.ulcerans_Agy99 ARSITIFAAASLKTAFTQIGERFKTANPG--VGVELDFAGSSELATQLTQ
MAV_2863|M.avium_104 AGKLMVFAAASLRPAFTQIAERFKAQNPG--TGIEFEFAGSSELATQLTQ
Mflv_3859|M.gilvum_PYR-GCK TGDLTVFAAASLKSTFTELGSRFETDHPG--TQVVFNFAGSTDLAAQISQ
Mvan_2538|M.vanbaalenii_PYR-1 NGTVTVFAASSLKNTFTELGAQFEKDHPG--TEVTFNFAGSADLVAQLSQ
MSMEG_2016|M.smegmatis_MC2_155 DDHLTVFAAASLKSVFTELGKRFEDANPG--ATVEFNFAGSSDLATQLTQ
MAB_2433|M.abscessus_ATCC_1997 PVRLHVLAAASLRKAFTEIGRSFESTHAG--TTVEFTFAGSSDLVTQLTQ
TH_1176|M.thermoresistible__bu STAVTVYAAASLQQAFTTLGELFEAEHPD--VTVRFTFAGSADLATQLLG
MLBr_00615|M.leprae_Br4923 HTSIALVAYSAPGPGWGKVIPAFNDARSGDGVQVVTSYGASGDQSRGVVD
: : * :: : : *: ... . : :..* : :
Mb1888|M.bovis_AF2122/97 GATADVFASADTAQMDSVAKAGLLAGHPTNFATN------TMVIVAAAGN
Rv1857|M.tuberculosis_H37Rv GATADVFASADTAQMDSVAKAGLLAGHPTNFATN------TMVIVAAAGN
MMAR_2731|M.marinum_M GATADVFASADTAQMDTVAKAGLLAADPTNFASN------TLVIVTAPGN
MUL_3022|M.ulcerans_Agy99 GATADVFASADTAQMDTVAKAGLLAADPTNFASN------TLVIVTAPGN
MAV_2863|M.avium_104 GATADVFASADTAQMDVVAKAGLLAADPTNFASN------TLVIVTAPGN
Mflv_3859|M.gilvum_PYR-GCK GAPADVFASADTSTMDRAVDGGTVAGDPVVFATN------TLTIVTALGN
Mvan_2538|M.vanbaalenii_PYR-1 GAPADVFASADTRNMTRAVDGGLVAGQPADFASN------TLTIVTPPGN
MSMEG_2016|M.smegmatis_MC2_155 GAPADVFASADAPNMNKVVDAGLVHGDPVDFAAN------TLTIVTAPGN
MAB_2433|M.abscessus_ATCC_1997 GAQADVLATADSANMDKAWKAHAVVN-PVDFAAN------TLTIVVAPGN
TH_1176|M.thermoresistible__bu GADADVFAAADEATMERVAAAGLLSGPPVIFAAN------TLTVVTAPGN
MLBr_00615|M.leprae_Br4923 GKPADIVNFSVETDITRLVKVGKVSKDWDTDATKGIPFGSVVTLVVRTGN
* **:. : : : *:: .:.:*. **
Mb1888|M.bovis_AF2122/97 PKKIRSFADLTRPGLNVVVCQPSVP-----------------CGSATRRI
Rv1857|M.tuberculosis_H37Rv PKKIRSFADLTRPGLNVVVCQPSVP-----------------CGSATRRI
MMAR_2731|M.marinum_M PKHVGSFADLSRSDLTVVVCQQQVP-----------------CGAATQRI
MUL_3022|M.ulcerans_Agy99 PKHVGSFADLSRSDLIVVVCQQQVP-----------------CGAATQRI
MAV_2863|M.avium_104 PKRIGSFADLARPGLTVVTCQRPVP-----------------CGAAAHRV
Mflv_3859|M.gilvum_PYR-GCK PRAIASFADLAGPGVQLVVCAPQVP-----------------CGSATEKV
Mvan_2538|M.vanbaalenii_PYR-1 PRGITSFGDLTRPGTQVVVCAPQVP-----------------CGSAAERV
MSMEG_2016|M.smegmatis_MC2_155 PKHITSFADLTKPGISVVVCAPQVP-----------------CGAATEKV
MAB_2433|M.abscessus_ATCC_1997 PKGIGAFGDLTRPGLDVVVCAPQVP-----------------CGAATAGV
TH_1176|M.thermoresistible__bu PHGLTGFADLARAGPAVVTCAPQVP-----------------CGAATQRL
MLBr_00615|M.leprae_Br4923 PKNIKDWDDLVRPGVEVIMPNPLSSGSAKWNLLAPYAAKSEGCRNSQAGI
*: : : ** .. :: . * : :
Mb1888|M.bovis_AF2122/97 EDATGIHLNPVSEEL-SVTDVLNKVITGQADAGLVYVSDALSVATKVTCV
Rv1857|M.tuberculosis_H37Rv EDATGIHLNPVSEEL-SVTDVLNKVITGQADAGLVYVSDALSVATKVTCV
MMAR_2731|M.marinum_M EDGTGVHLNPVSEEL-SVTDVLNKVTTGQADAALVYLTDAMSAGSTVSTI
MUL_3022|M.ulcerans_Agy99 EDGTGVHLNRVSEEL-SVTDVLNKVTTGQADAALVYLTDAMSAGSTVSTI
MAV_2863|M.avium_104 EDSTGVHLNPVSEEP-SVTDALTKVTSGQADAALVYVTDARTAGSKVATV
Mflv_3859|M.gilvum_PYR-GCK EAATGVTLTPVSEES-AVTDVLGKVTSGQADAGLVYVTDAAAAGDAVTAV
Mvan_2538|M.vanbaalenii_PYR-1 EEAAGATLTPVSEES-AVADVLGKVTSGQADAGLVYVTDAVAAGDRVTEI
MSMEG_2016|M.smegmatis_MC2_155 EKATGVTLHPVSEES-AVTDVLGKVTSGQADAGLVYVTDAKGAGAAVAAV
MAB_2433|M.abscessus_ATCC_1997 SKETGVHLNPASEES-SVTDVLNKVIAGQADAGLVYVTDAKAAADKVATV
TH_1176|M.thermoresistible__bu EQLTGVDLAPVSEEN-SVSDVLHKVTVGQADAGVVYRTDARRAGDTVHTV
MLBr_00615|M.leprae_Br4923 DFVNRLVAEHVKLRLGSAREATDAFVQGSGDVLISYENEAIAAERQGKPV
. .. . :. :. . *..*. : * .:* . :
Mb1888|M.bovis_AF2122/97 RFPEAAGVVNVY-AIAVLKRTSQPALARQFVAMVTAAAGRRILDQSGFAK
Rv1857|M.tuberculosis_H37Rv RFPEAAGVVNVY-AIAVLKRTSQPALARQFVAMVTAAAGRRILDQSGFAK
MMAR_2731|M.marinum_M KFPQAAPAVNVY-PIAVLSNAPQPGLARKFVDMVTAETGRRILDQSGFAK
MUL_3022|M.ulcerans_Agy99 KFPQAAPAVNVY-PIAVLSNAPQPGLARKFVDMVTAETGRRILDQSGFAK
MAV_2863|M.avium_104 NFPEAAGAVNVY-PIGVLKQAPLATQARNFVDLVTSPPGQQILAQAGFAK
Mflv_3859|M.gilvum_PYR-GCK AFPEAADAVNSY-PIAALADSTNPAAASEFIEFVTGPQGRDVLAAAGFGR
Mvan_2538|M.vanbaalenii_PYR-1 AFPESGVAANTY-PIAALAGSRNPDAAQQFMALVTGPQGREVLSAAGFGP
MSMEG_2016|M.smegmatis_MC2_155 AFGESADAVNTY-PIATLKDSGDIETAQRFVDLVVSPEGQEVLSEAGFVA
MAB_2433|M.abscessus_ATCC_1997 EIPEARSIVNTY-PIAATANSRHPERAAQFIETVTGDIGHKVLAAAGFSA
TH_1176|M.thermoresistible__bu EVPAAAAVPNRY-PVAVPADAARPDLGQRFIDLITGAAGRDVLAQNGFSE
MLBr_00615|M.leprae_Br4923 EHINPPVTFKIENPLAVVSTSAHLEVATAFKNFQYTTAAQGLWAQAGFRP
. : .:.. : . * .: : **
Mb1888|M.bovis_AF2122/97 P-------------------------------------------------
Rv1857|M.tuberculosis_H37Rv P-------------------------------------------------
MMAR_2731|M.marinum_M P-------------------------------------------------
MUL_3022|M.ulcerans_Agy99 P-------------------------------------------------
MAV_2863|M.avium_104 P-------------------------------------------------
Mflv_3859|M.gilvum_PYR-GCK P-------------------------------------------------
Mvan_2538|M.vanbaalenii_PYR-1 P-------------------------------------------------
MSMEG_2016|M.smegmatis_MC2_155 P-------------------------------------------------
MAB_2433|M.abscessus_ATCC_1997 P-------------------------------------------------
TH_1176|M.thermoresistible__bu PR------------------------------------------------
MLBr_00615|M.leprae_Br4923 VDPAVSADFSDQFPAPVKLWTIADLGGWSTVDPQLFDKNTGSITKVYMQA
Mb1888|M.bovis_AF2122/97 --
Rv1857|M.tuberculosis_H37Rv --
MMAR_2731|M.marinum_M --
MUL_3022|M.ulcerans_Agy99 --
MAV_2863|M.avium_104 --
Mflv_3859|M.gilvum_PYR-GCK --
Mvan_2538|M.vanbaalenii_PYR-1 --
MSMEG_2016|M.smegmatis_MC2_155 --
MAB_2433|M.abscessus_ATCC_1997 --
TH_1176|M.thermoresistible__bu --
MLBr_00615|M.leprae_Br4923 TG