For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGCGSPSRPAEGARSITIFAAASLKTAFTQIGERFKTANPGVGVELDFAGSSELATQLTQGATADVFASA DTAQMDTVAKAGLLAADPTNFASNTLVIVTAPGNPKHVGSFADLSRSDLIVVVCQQQVPCGAATQRIEDG TGVHLNRVSEELSVTDVLNKVTTGQADAALVYLTDAMSAGSTVSTIKFPQAAPAVNVYPIAVLSNAPQPG LARKFVDMVTAETGRRILDQSGFAKP*
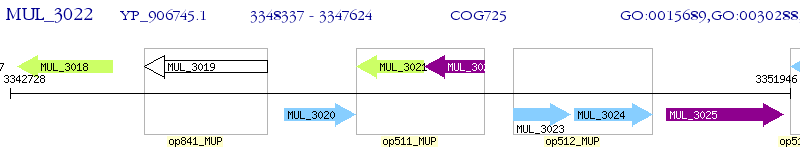
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3022 | modA | - | 100% (237) | molybdate-binding lipoprotein ModA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1888 | modA | 8e-93 | 71.19% (243) | molybdate-binding lipoprotein |
| M. gilvum PYR-GCK | Mflv_3859 | - | 2e-67 | 55.66% (221) | molybdenum ABC transporter, periplasmic molybdate-binding |
| M. tuberculosis H37Rv | Rv1857 | modA | 8e-93 | 71.19% (243) | molybdate-binding lipoprotein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2433 | - | 8e-65 | 53.62% (235) | molybdenum ABC transporter periplasmic protein |
| M. marinum M | MMAR_2731 | modA | 1e-128 | 98.73% (237) | molybdate-binding lipoprotein ModA |
| M. avium 104 | MAV_2863 | modA | 4e-94 | 72.57% (237) | molybdate ABC transporter, periplasmic molybdate-binding |
| M. smegmatis MC2 155 | MSMEG_2016 | modA | 2e-72 | 59.28% (221) | molybdate ABC transporter, periplasmic molybdate-binding |
| M. thermoresistible (build 8) | TH_1176 | - | 2e-67 | 54.85% (237) | PROBABLE MOLYBDATE-BINDING LIPOPROTEIN MODA |
| M. vanbaalenii PYR-1 | Mvan_2538 | - | 4e-64 | 51.25% (240) | molybdenum ABC transporter, periplasmic molybdate-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3022|M.ulcerans_Agy99 ------------------------------MVG-CGS------PSRPAEG
MMAR_2731|M.marinum_M -MTRVG-----------MLAGMVSALLVW-VVG-CGS------PSRPADG
Mb1888|M.bovis_AF2122/97 -MRWIG-----------LSTGLVSAMLVAGLVA-CGSNSPASSPAGPTQG
Rv1857|M.tuberculosis_H37Rv -MRWIG-----------LSTGLVSAMLVAGLVA-CGSNSPASSPAGPTQG
MAV_2863|M.avium_104 -MRRIG-----------ILTGLLSVVLIAGMTG-CGS------KSQPPPT
Mflv_3859|M.gilvum_PYR-GCK MIRHPA-----------VAVALTVATLMSGCST-AGEDRPAATPPTSDAV
Mvan_2538|M.vanbaalenii_PYR-1 MRSMPG-----------LTSALIAVVLVAGCSS-SRTD-SGPTP------
MSMEG_2016|M.smegmatis_MC2_155 ---MIR-----------VVSVVVGVFLLAGCTP-APAN------------
MAB_2433|M.abscessus_ATCC_1997 MTGSTRNVLTSRAWKVLAATLLAGSLIVSGCGTRTGSGGGSASPSTGAGD
TH_1176|M.thermoresistible__bu MIRVRR------------WLSVCLALMLPAVAG-CGTG--------RTGG
MUL_3022|M.ulcerans_Agy99 ARSITIFAAASLKTAFTQIGERFKTANPGVGVELDFAGSSELATQLTQGA
MMAR_2731|M.marinum_M ARSITIFAAASLKTAFTQIGERFKTANPGVGVELDFAGSSELATQLTQGA
Mb1888|M.bovis_AF2122/97 ARSIVVFAAASLQSAFTQIGEQFKAGNPGVNVNFAFAGSSELATQLTQGA
Rv1857|M.tuberculosis_H37Rv ARSIVVFAAASLQSAFTQIGEQFKAGNPGVNVNFAFAGSSELATQLTQGA
MAV_2863|M.avium_104 AGKLMVFAAASLRPAFTQIAERFKAQNPGTGIEFEFAGSSELATQLTQGA
Mflv_3859|M.gilvum_PYR-GCK TGDLTVFAAASLKSTFTELGSRFETDHPGTQVVFNFAGSTDLAAQISQGA
Mvan_2538|M.vanbaalenii_PYR-1 NGTVTVFAASSLKNTFTELGAQFEKDHPGTEVTFNFAGSADLVAQLSQGA
MSMEG_2016|M.smegmatis_MC2_155 DDHLTVFAAASLKSVFTELGKRFEDANPGATVEFNFAGSSDLATQLTQGA
MAB_2433|M.abscessus_ATCC_1997 PVRLHVLAAASLRKAFTEIGRSFESTHAGTTVEFTFAGSSDLVTQLTQGA
TH_1176|M.thermoresistible__bu STAVTVYAAASLQQAFTTLGELFEAEHPDVTVRFTFAGSADLATQLLGGA
: : **:**: .** :. *: :... : : ****::*.:*: **
MUL_3022|M.ulcerans_Agy99 TADVFASADTAQMDTVAKAGLLAADPTNFASNTLVIVTAPGNPKHVGSFA
MMAR_2731|M.marinum_M TADVFASADTAQMDTVAKAGLLAADPTNFASNTLVIVTAPGNPKHVGSFA
Mb1888|M.bovis_AF2122/97 TADVFASADTAQMDSVAKAGLLAGHPTNFATNTMVIVAAAGNPKKIRSFA
Rv1857|M.tuberculosis_H37Rv TADVFASADTAQMDSVAKAGLLAGHPTNFATNTMVIVAAAGNPKKIRSFA
MAV_2863|M.avium_104 TADVFASADTAQMDVVAKAGLLAADPTNFASNTLVIVTAPGNPKRIGSFA
Mflv_3859|M.gilvum_PYR-GCK PADVFASADTSTMDRAVDGGTVAGDPVVFATNTLTIVTALGNPRAIASFA
Mvan_2538|M.vanbaalenii_PYR-1 PADVFASADTRNMTRAVDGGLVAGQPADFASNTLTIVTPPGNPRGITSFG
MSMEG_2016|M.smegmatis_MC2_155 PADVFASADAPNMNKVVDAGLVHGDPVDFAANTLTIVTAPGNPKHITSFA
MAB_2433|M.abscessus_ATCC_1997 QADVLATADSANMDKAWKAHAVVN-PVDFAANTLTIVVAPGNPKGIGAFG
TH_1176|M.thermoresistible__bu DADVFAAADEATMERVAAAGLLSGPPVIFAANTLTVVTAPGNPHGLTGFA
***:*:** * . . : *. **:**:.:*.. ***: : .*.
MUL_3022|M.ulcerans_Agy99 DLSRSDLIVVVCQQQVPCGAATQRIEDGTGVHLNRVSEELSVTDVLNKVT
MMAR_2731|M.marinum_M DLSRSDLTVVVCQQQVPCGAATQRIEDGTGVHLNPVSEELSVTDVLNKVT
Mb1888|M.bovis_AF2122/97 DLTRPGLNVVVCQPSVPCGSATRRIEDATGIHLNPVSEELSVTDVLNKVI
Rv1857|M.tuberculosis_H37Rv DLTRPGLNVVVCQPSVPCGSATRRIEDATGIHLNPVSEELSVTDVLNKVI
MAV_2863|M.avium_104 DLARPGLTVVTCQRPVPCGAAAHRVEDSTGVHLNPVSEEPSVTDALTKVT
Mflv_3859|M.gilvum_PYR-GCK DLAGPGVQLVVCAPQVPCGSATEKVEAATGVTLTPVSEESAVTDVLGKVT
Mvan_2538|M.vanbaalenii_PYR-1 DLTRPGTQVVVCAPQVPCGSAAERVEEAAGATLTPVSEESAVADVLGKVT
MSMEG_2016|M.smegmatis_MC2_155 DLTKPGISVVVCAPQVPCGAATEKVEKATGVTLHPVSEESAVTDVLGKVT
MAB_2433|M.abscessus_ATCC_1997 DLTRPGLDVVVCAPQVPCGAATAGVSKETGVHLNPASEESSVTDVLNKVI
TH_1176|M.thermoresistible__bu DLARAGPAVVTCAPQVPCGAATQRLEQLTGVDLAPVSEENSVSDVLHKVT
**: .. :*.* ****:*: :. :* * .*** :*:*.* **
MUL_3022|M.ulcerans_Agy99 TGQADAALVYLTDAMSAGSTVSTIKFPQAAPAVNVYPIAVLSNAPQPGLA
MMAR_2731|M.marinum_M TGQADAALVYLTDAMSAGSTVSTIKFPQAAPAVNVYPIAVLSNAPQPGLA
Mb1888|M.bovis_AF2122/97 TGQADAGLVYVSDALSVATKVTCVRFPEAAGVVNVYAIAVLKRTSQPALA
Rv1857|M.tuberculosis_H37Rv TGQADAGLVYVSDALSVATKVTCVRFPEAAGVVNVYAIAVLKRTSQPALA
MAV_2863|M.avium_104 SGQADAALVYVTDARTAGSKVATVNFPEAAGAVNVYPIGVLKQAPLATQA
Mflv_3859|M.gilvum_PYR-GCK SGQADAGLVYVTDAAAAGDAVTAVAFPEAADAVNSYPIAALADSTNPAAA
Mvan_2538|M.vanbaalenii_PYR-1 SGQADAGLVYVTDAVAAGDRVTEIAFPESGVAANTYPIAALAGSRNPDAA
MSMEG_2016|M.smegmatis_MC2_155 SGQADAGLVYVTDAKGAGAAVAAVAFGESADAVNTYPIATLKDSGDIETA
MAB_2433|M.abscessus_ATCC_1997 AGQADAGLVYVTDAKAAADKVATVEIPEARSIVNTYPIAATANSRHPERA
TH_1176|M.thermoresistible__bu VGQADAGVVYRTDARRAGDTVHTVEVPAAAAVPNRYPVAVPADAARPDLG
*****.:** :** .. * : . : * *.:.. : .
MUL_3022|M.ulcerans_Agy99 RKFVDMVTAETGRRILDQSGFAKP-
MMAR_2731|M.marinum_M RKFVDMVTAETGRRILDQSGFAKP-
Mb1888|M.bovis_AF2122/97 RQFVAMVTAAAGRRILDQSGFAKP-
Rv1857|M.tuberculosis_H37Rv RQFVAMVTAAAGRRILDQSGFAKP-
MAV_2863|M.avium_104 RNFVDLVTSPPGQQILAQAGFAKP-
Mflv_3859|M.gilvum_PYR-GCK SEFIEFVTGPQGRDVLAAAGFGRP-
Mvan_2538|M.vanbaalenii_PYR-1 QQFMALVTGPQGREVLSAAGFGPP-
MSMEG_2016|M.smegmatis_MC2_155 QRFVDLVVSPEGQEVLSEAGFVAP-
MAB_2433|M.abscessus_ATCC_1997 AQFIETVTGDIGHKVLAAAGFSAP-
TH_1176|M.thermoresistible__bu QRFIDLITGAAGRDVLAQNGFSEPR
.*: :.. *: :* ** *