For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQPYGQYCPVARAAELLGDRWTLLIVRELLFGPLRFTEIERGLPGISRSVLAQRLRRLQHDRIIEAVPEH TGGGYRFTVAGEELRPVLQTLGDWVSRWLMADPTPAECDPELLTLWISRRVNTEALPGRRVVVEFRYHGE RPLWAWLVLEPGDISVCLHDPCLPVDLTVRGHPRDLYRVYSGRSTLAAEISAERIELDGLPAMRRAFPSW MAWSPFAPAMRQAVVSVDQMPEAHGG
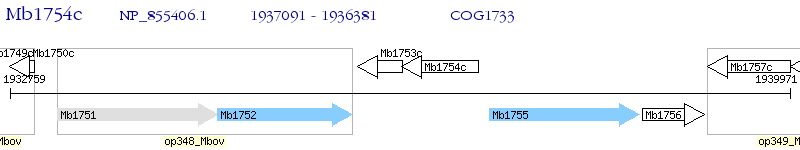
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1754c | - | - | 100% (236) | hypothetical protein Mb1754c |
| M. bovis AF2122 / 97 | Mb3122 | - | 1e-11 | 41.94% (93) | putative transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5238 | - | 1e-19 | 31.68% (202) | HxlR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1725c | - | 1e-140 | 100.00% (236) | hypothetical protein Rv1725c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3456 | - | 5e-12 | 35.24% (105) | HxlR family transcriptional regulator |
| M. marinum M | MMAR_1626 | - | 1e-33 | 36.04% (222) | transcriptional regulator |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5032 | - | 1e-14 | 41.18% (102) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_2405 | - | 6e-15 | 31.31% (214) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_3410 | - | 6e-34 | 36.04% (222) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1032 | - | 4e-21 | 34.15% (205) | HxlR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5032|M.smegmatis_MC2_155 ------MASRSYGQYCALAKSLDLVGDRWTLLVVRELLDGPRRYGDLLSA
TH_2405|M.thermoresistible__bu ------MASRTYGQHCALAKSLDLVGDRWTLLIVRELLDGPRRYSDLLTA
Mflv_5238|M.gilvum_PYR-GCK MGRLADVSRRTYGQYCGLAHALDVVGERWTLLIVRELASGPKRYTELADA
Mvan_1032|M.vanbaalenii_PYR-1 MSTIVGMTRRSYGQFCGLAHALDVVGERWTLLIVRELGSGPKRYGELADA
Mb1754c|M.bovis_AF2122/97 --------MQPYGQYCPVARAAELLGDRWTLLIVRELLFGPLRFTEIERG
Rv1725c|M.tuberculosis_H37Rv --------MQPYGQYCPVARAAELLGDRWTLLIVRELLFGPLRFTEIERG
MMAR_1626|M.marinum_M --------MSTYSQFCPVAKAMELLDERWTLLVVRELLLGSTHFNDLRRG
MUL_3410|M.ulcerans_Agy99 --------MSTYSQFCPVAKAMELLDERWTLLVVRELLLGSTHFNDLRRG
MAB_3456|M.abscessus_ATCC_1997 -------MDFTTMRNCSISNALDVIGERWTLVALREIMLGNRRFETIVRN
. : * ::.: :::.:****: :**: * :: :
MSMEG_5032|M.smegmatis_MC2_155 LSPIATDVLAGRLRDLEANNLVHKR-AMPRPSATTVYALTADGRALEDVI
TH_2405|M.thermoresistible__bu LRPIATDMLATRLRHLEEHHLVRRR-ELPKPASGTVYELTADGAALEAVV
Mflv_5238|M.gilvum_PYR-GCK LAGIGTSLLATRVRQLETDGVVQRRLALDQPGSTVVYELSDAGRELAAAV
Mvan_1032|M.vanbaalenii_PYR-1 LAGIGTSLLATRMRQLETDGVIQRRLDLDQPGSAVVYELSDAGRELAEAM
Mb1754c|M.bovis_AF2122/97 LPGISRSVLAQRLRRLQHDRIIEAV----PEHTGGGYRFTVAGEELRPVL
Rv1725c|M.tuberculosis_H37Rv LPGISRSVLAQRLRRLQHDRIIEAV----PEHTGGGYRFTVAGEELRPVL
MMAR_1626|M.marinum_M VPKMSPALLSKRLKSLTKAGVVERS----DIEGRTSYSLTTCGRELATVV
MUL_3410|M.ulcerans_Agy99 VPKMSPALLSKRLKSLTKAAVVERS----DIEGRTSYSLTTCGRELATVV
MAB_3456|M.abscessus_ATCC_1997 TG-ASRDILATRLRKLVDAGVLEKR-RYEDRPPRYEYLLTQSGQALRPVL
. :*: *:: * ::. * :: * * .:
MSMEG_5032|M.smegmatis_MC2_155 NAFARWGR--HLLANRDRKETVRPEWLVRAVRAYVRDD-RTGPPVTVRMV
TH_2405|M.thermoresistible__bu HAFIRWGR--SLLETRRAGDVVRPHWLARAVRAHVRPD-RTGPPLSVRLV
Mflv_5238|M.gilvum_PYR-GCK VPLAMWGARHQMADADVCHEAFRAEWLLSFLAADVRRDGPHDLDAVYEFH
Mvan_1032|M.vanbaalenii_PYR-1 IPLAMWGARHQMSHADPCAEAFRAEWLLAFLAADLRGGIPEGLASVYEFH
Mb1754c|M.bovis_AF2122/97 QTLGDWVS--RWLMADPTPAECDPELLTLWISRRVNTEALPGRRVVVEFR
Rv1725c|M.tuberculosis_H37Rv QTLGDWVS--RWLMADPTPAECDPELLTLWISRRVNTEALPGRRVVVEFR
MMAR_1626|M.marinum_M DALGEWGI--RWIP-ELGDEDLDPHLLMWDIRRTVPIDAWPRARTVLAFR
MUL_3410|M.ulcerans_Agy99 DALGEWGI--RWIP-ELGDEDLDPHLLMWDIRRTVPIDAWPRARTVLAFR
MAB_3456|M.abscessus_ATCC_1997 FALMEWGD-----------------------------------KFVTKG-
.: *
MSMEG_5032|M.smegmatis_MC2_155 TPE-----GQAAVEINEHTVESLDD--EADVDVTLTGAAEVLAAAMDPG-
TH_2405|M.thermoresistible__bu TPE-----GAAAVRISADAVEDLDD--DTAVDVTLTGAAEVLAAALHPD-
Mflv_5238|M.gilvum_PYR-GCK IDD-----STACLRLRDGGVTVTPGAPATPGDVVVRSRASTIAEIVGHKI
Mvan_1032|M.vanbaalenii_PYR-1 VDD-----STACLQIRDGRVSVRPGPCATPADVTVRATARAIAAVVGQKD
Mb1754c|M.bovis_AF2122/97 YHGERPLWA--WLVLEPGDISVCLHDPCLPVDLTVRGHPRDLYRVYSGRS
Rv1725c|M.tuberculosis_H37Rv YHGERPLWA--WLVLEPGDISVCLHDPCLPVDLTVRGHPRDLYRVYSGRS
MMAR_1626|M.marinum_M LAGVAPKAARWWLLVSEGEADVCDFDPGYQVAATVETSLLTLTQLWRGDQ
MUL_3410|M.ulcerans_Agy99 LAGVAPKAARWWLLVSEGEADVCDFDPGYQVAATVETSLLTLTQLWRGDQ
MAB_3456|M.abscessus_ATCC_1997 -----------------------------PPPSIWEHQCGSVLHIQPTCE
:
MSMEG_5032|M.smegmatis_MC2_155 QVENLVAAGLLTIDGDARAVRRLAKAFAPPRVRV----------------
TH_2405|M.thermoresistible__bu RAAELVAQGRLRVDGRADALRRLAEVVTAPAGRAVPDRS-----------
Mflv_5238|M.gilvum_PYR-GCK AVADALADGSLEITGDADKISALVGVIEKRLAR-----------------
Mvan_1032|M.vanbaalenii_PYR-1 RLEDAVAEGRIEVTGDPAAIATLIGAVEKRLA------------------
Mb1754c|M.bovis_AF2122/97 TLAAEISAERIELDGLPAMRRAFPSWMAWSPFAPAMRQAVVSVDQMPEAH
Rv1725c|M.tuberculosis_H37Rv TLAAEISAERIELDGLPAMRRAFPSWMAWSPFAPAMRQAVVSVDQMPEAH
MMAR_1626|M.marinum_M SWSRAMLNGGVRIDGPAQVRRSVPAWLGQSRLAAVPRPA-----------
MUL_3410|M.ulcerans_Agy99 SWSRAMLNGGVRIDGPAQVRRSVPAWLGQSRLAAVPRPA-----------
MAB_3456|M.abscessus_ATCC_1997 SCGEAVTFDDLTPRRLGRVR------------------------------
: :
MSMEG_5032|M.smegmatis_MC2_155 --
TH_2405|M.thermoresistible__bu --
Mflv_5238|M.gilvum_PYR-GCK --
Mvan_1032|M.vanbaalenii_PYR-1 --
Mb1754c|M.bovis_AF2122/97 GG
Rv1725c|M.tuberculosis_H37Rv GG
MMAR_1626|M.marinum_M --
MUL_3410|M.ulcerans_Agy99 --
MAB_3456|M.abscessus_ATCC_1997 --