For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MMDRIVMEVSRYRPEIESAPTFQAYEVPLTREWAVLDGLTYIKDHLDGTLSFRWSCRMGICGSSGMTING DPKLACATFLADYLPGPVRVEPMRNFPVIRDLVVDISDFMAKLPSVKPWLVRHDEPPVEDGEYRQTPAEL DAFKQFSMCINCMLCYSACPVYALDPDFLGPAAIALGQRYNLDSRDQGAADRRDVLAAADGAWACTLVGE CSTACPKGVDPAGAIQRYKLTAATHALKKLLFPWGGGRMSAYRQPVERYWWARRRSYLRFMLREISCIFV AWFVLYLVLVLRAVGAGGNSYQRFLDFSANPVVVVLNVVALSFLLLHAVTWFGSAPRAMVIQVRGRRVPA RAVLAGHYAAWLVVSVIVAWMVLS
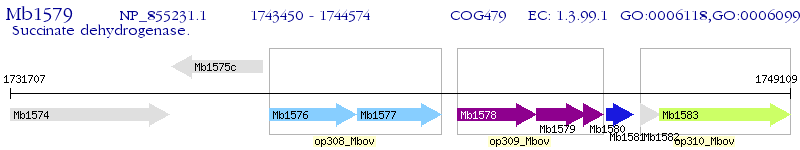
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1579 | frdBC | - | 100% (374) | fumarate reductase iron-sulfur subunit FrdB/membrane anchor subunit FrdC |
| M. bovis AF2122 / 97 | Mb3348 | sdhB | 9e-44 | 37.87% (235) | succinate dehydrogenase iron-sulfur subunit |
| M. bovis AF2122 / 97 | Mb0253c | - | 3e-21 | 31.31% (214) | fumarate reductase iron-sulfur subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4847 | sdhB | 2e-43 | 37.66% (231) | succinate dehydrogenase iron-sulfur subunit |
| M. tuberculosis H37Rv | Rv1553 | frdB | 1e-148 | 100.00% (247) | fumarate reductase iron-sulfur subunit FrdB |
| M. leprae Br4923 | MLBr_00696 | sdhB | 3e-42 | 36.60% (235) | succinate dehydrogenase iron-sulfur subunit |
| M. abscessus ATCC 19977 | MAB_3676 | sdhB | 2e-44 | 38.43% (229) | succinate dehydrogenase iron-sulfur subunit |
| M. marinum M | MMAR_1200 | sdhB | 5e-44 | 38.30% (235) | succinate dehydrogenase (iron-sulphur protein subunit) SdhB |
| M. avium 104 | MAV_4300 | sdhB | 2e-45 | 38.49% (239) | succinate dehydrogenase iron-sulfur subunit |
| M. smegmatis MC2 155 | MSMEG_1669 | sdhB | 2e-45 | 38.96% (231) | succinate dehydrogenase iron-sulfur subunit |
| M. thermoresistible (build 8) | TH_2183 | sdhB | 6e-46 | 39.39% (231) | PROBABLE SUCCINATE DEHYDROGENASE (IRON-SULPHUR PROTEIN |
| M. ulcerans Agy99 | MUL_1370 | sdhB | 4e-45 | 39.15% (235) | succinate dehydrogenase iron-sulfur subunit |
| M. vanbaalenii PYR-1 | Mvan_1586 | sdhB | 6e-44 | 37.66% (231) | succinate dehydrogenase iron-sulfur subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4847|M.gilvum_PYR-GCK ---MSAPV--LDKQ-----DTPPVPEGAVMVTLKIARFNPESPDD----A
Mvan_1586|M.vanbaalenii_PYR-1 ---MSAPT--LEKA-----ELPPVPEGAVMVTLKIARFNPESPDD----A
MSMEG_1669|M.smegmatis_MC2_155 ---MSAPV--IDKPEAGDPELPPVPEGAVMVTLKIARFNPENPDA----A
TH_2183|M.thermoresistible__bu --MTITPD--IDKPATTDAPLPPVPEGAVMVTLKIARFNPENPDA----A
MAB_3676|M.abscessus_ATCC_1997 ----MTAV--IEKAEAGDPPLPPVPEGAVMVDLKIQRFNPEDPDA----A
MLBr_00696|M.leprae_Br4923 --MAVEPV----EVKSLEPPRPPISDDAVMVTVKIARFNPDNPDAFAETG
MAV_4300|M.avium_104 --MSVEP-----DVETLEPPLPPVPDEAVMVTVKIARFNPDQPDAYAETG
MMAR_1200|M.marinum_M MSLDTERASSDDVTDSLAPPLPPVPDGAVMVTVKIARFNPDDPEAFASTG
MUL_1370|M.ulcerans_Agy99 --MDTERASSDDVTDSLAPPLPPVPDGAVMVTVKIARYNPDDPEAFASTG
Mb1579|M.bovis_AF2122/97 --------------------------MMDRIVMEVSRYRPEIESA----P
Rv1553|M.tuberculosis_H37Rv --------------------------MMDRIVMEVSRYRPEIESA----P
: ::: *:.*: .
Mflv_4847|M.gilvum_PYR-GCK GWQSFRVPCLPSDRLLNLLHYVKWYLDGTLTFRRSCAHGVCGSDAMRING
Mvan_1586|M.vanbaalenii_PYR-1 GWQSFRVPCLPSDRLLNLLHYVKWYLDGTLTFRRSCAHGVCGSDAMRING
MSMEG_1669|M.smegmatis_MC2_155 GWQSFRVPCLPSDRLLNLLHYVKWYLDGTLTFRRSCAHGVCGSDAMRING
TH_2183|M.thermoresistible__bu GFQSFRVPTLPTDRLLNLLHYVKWYLDGTLTFRRSCAHGVCGSDAMRVNG
MAB_3676|M.abscessus_ATCC_1997 GWQTFRVPCLPTDRLLNLLLYVKGYLDGTLTFRRSCAHGVCGSDAMRING
MLBr_00696|M.leprae_Br4923 GWQSFRVPCLPTDRLLNLLIYIKGYLDGTLTFRRSCAHGVCGSDAMRVNG
MAV_4300|M.avium_104 GWQSFRVPCLPSDRMLNLLIYIKSYLDGTLTFRRSCAHGVCGSDAMRING
MMAR_1200|M.marinum_M GWQSFRVPCLPSDRLLNLLIYIKGYLDGTLTFRRSCAHGVCGSDAMRING
MUL_1370|M.ulcerans_Agy99 GWQSFRVPCLPSDRLLNLLIYIKGYLDGTLTFRRSCAHGVCGSDAMRING
Mb1579|M.bovis_AF2122/97 TFQAYEVPLTREWAVLDGLTYIKDHLDGTLSFRWSCRMGICGSSGMTING
Rv1553|M.tuberculosis_H37Rv TFQAYEVPLTREWAVLDGLTYIKDHLDGTLSFRWSCRMGICGSSGMTING
:*::.** :*: * *:* :*****:** ** *:***..* :**
Mflv_4847|M.gilvum_PYR-GCK VNRLACKVLMRDMLPK----NPNKQLTITIEPIRGLPVEKDLVVDMEPFF
Mvan_1586|M.vanbaalenii_PYR-1 VNRLACKVLMRDMLPK----NPKKQLTITIEPIRGLPVEKDLVVDMEPFF
MSMEG_1669|M.smegmatis_MC2_155 VNRLACKVLMRDMLPK----NPNKQLTITIEPIRGLPVEKDLVVNMEPFF
TH_2183|M.thermoresistible__bu VNRLACKVLMRDLLPK----NPKKELTVTVEPIRGLPVEKDLVVDMEPFF
MAB_3676|M.abscessus_ATCC_1997 VNRLACKVLMRDLLPK----NPKKTLTITIEPIRGLPVEKDLVVNMEPFF
MLBr_00696|M.leprae_Br4923 VNRLACKVLMRDLLPK----KAGKNLTITVEPIRGLPVEKDLVVDMEPFF
MAV_4300|M.avium_104 VNRLACKVLMRDLLPEPKKGKPGKPLTITVEPIRGLPVEKDLVVDMEPFF
MMAR_1200|M.marinum_M VNRLACKVLMRDLLPK----KPGKQLTLTVEPIRGLPVEKDLVVDMEPFF
MUL_1370|M.ulcerans_Agy99 VNRLACKVLMRDLLPK----KSGKQLTLTVEPIRGLPVEKDLVVDMEPFF
Mb1579|M.bovis_AF2122/97 DPKLACATFLADYLPG----------PVRVEPMRNFPVIRDLVVDISDFM
Rv1553|M.tuberculosis_H37Rv DPKLACATFLADYLPG----------PVRVEPMRNFPVIRDLVVDISDFM
:*** .:: * ** .: :**:*.:** :****::. *:
Mflv_4847|M.gilvum_PYR-GCK DAFRAVKPYLITSGNQPTR--ERIQSQTDRARYDDTTKCILCACCTTSCP
Mvan_1586|M.vanbaalenii_PYR-1 DAFRAIKPYLITSGNQPTR--ERIQSQTDRARYDDTTKCILCACCTTSCP
MSMEG_1669|M.smegmatis_MC2_155 DAYRAVKPFLVTSGNPPTK--ERIQSPTDRARYDDTTKCILCACCTTSCP
TH_2183|M.thermoresistible__bu DAYRAVKPFLVTSGNPPTR--ERLQSPQDRARYDDTTKCILCACCTTSCP
MAB_3676|M.abscessus_ATCC_1997 DAYRAVKPFMITSGNQPTR--EYIQSQTDRARFDDTTKCILCACCTTSCP
MLBr_00696|M.leprae_Br4923 DAYRAVKPYLITSGNPPTR--ERIQSPTDRARYDDTTKCILCACCTTSCP
MAV_4300|M.avium_104 DAYRAVKPYLITTGNPPTR--ERIQSPADRARYDDTTKCILCAACTTSCP
MMAR_1200|M.marinum_M DAYRAVKPYLITSGNPPTR--ERIQSPTDRARYDDTTKCILCACCTTSCP
MUL_1370|M.ulcerans_Agy99 DAYRAVKPYLITSGNPPTR--ERIQSPTDRARYDDTTKCILCACCTTSCP
Mb1579|M.bovis_AF2122/97 AKLPSVKPWLVRHDEPPVEDGEYRQTPAELDAFKQFSMCINCMLCYSACP
Rv1553|M.tuberculosis_H37Rv AKLPSVKPWLVRHDEPPVEDGEYRQTPAELDAFKQFSMCINCMLCYSACP
::**::: .: *.. * *: : :.: : ** * * ::**
Mflv_4847|M.gilvum_PYR-GCK VYWSEGSYFGPAAIVNAHRFIFDSRDEAAAERLDILNDIDGVWRCRTTFN
Mvan_1586|M.vanbaalenii_PYR-1 VYWNEGSYFGPAAIVNAHRFIFDSRDEGAAERLDILNDVDGVWRCRTTFN
MSMEG_1669|M.smegmatis_MC2_155 VYWSEGSYFGPAAIVNAHRFIFDSRDEAAAERLDILNEVDGVWRCRTTFN
TH_2183|M.thermoresistible__bu VYWTEGSYFGPAAIVNAHRFIFDSRDEAAGERLDILNEVDGVWRCRTTFN
MAB_3676|M.abscessus_ATCC_1997 VFWSDGSYFGPAAIVNAHRFIFDSRDEAAAERLDILNDVDGVWRCRTTFN
MLBr_00696|M.leprae_Br4923 VFWSEGTYYGPAAIVNAHRFIFDSRDEGAGERLTLLNEVDGVWRCRTTFN
MAV_4300|M.avium_104 IFWHEGSYYGPAAIVNAHRFIFDSRDEGAAERLDILNEVDGVWRCRTTFN
MMAR_1200|M.marinum_M VFWNEGSYFGPAAIVNAHRFIFDSRDEAAAERLDILNEVDGVWRCRTTFN
MUL_1370|M.ulcerans_Agy99 VFWNEGSYLGPAAIVNAHRFIFDSRDEAAAERVDILNEVDGVWRCRTTFN
Mb1579|M.bovis_AF2122/97 VYALDPDFLGPAAIALGQRYNLDSRDQGAADRRDVLAAADGAWACTLVGE
Rv1553|M.tuberculosis_H37Rv VYALDPDFLGPAAIALGQRYNLDSRDQGAADRRDVLAAADGAWACTLVGE
:: : : *****. .:*: :****:.*.:* :* **.* * . :
Mflv_4847|M.gilvum_PYR-GCK CTEACPRGIQVTKAIQEVK--------RALMFAR----------------
Mvan_1586|M.vanbaalenii_PYR-1 CTEACPRGIQVTKAIQEVK--------RALMFAR----------------
MSMEG_1669|M.smegmatis_MC2_155 CTEACPRGIQVTQAIQEVK--------RALMFAR----------------
TH_2183|M.thermoresistible__bu CTESCPRGIQVTKAIQEVK--------RALMFAR----------------
MAB_3676|M.abscessus_ATCC_1997 CTDACPRGIEVTKAIQEVK--------RALMFAR----------------
MLBr_00696|M.leprae_Br4923 CTESCPRNIEVTKAIQEVK--------RALMFSR----------------
MAV_4300|M.avium_104 CTDACPRGIEVTKAIQEVK--------RAIMFSR----------------
MMAR_1200|M.marinum_M CTEACPRGIEVTKAIQEVK--------RALLFSR----------------
MUL_1370|M.ulcerans_Agy99 CTEACPRGIEATKAIQEVK--------RALLFSR----------------
Mb1579|M.bovis_AF2122/97 CSTACPKGVDPAGAIQRYKLTAATHALKKLLFPWGGGRMSAYRQPVERYW
Rv1553|M.tuberculosis_H37Rv CSTACPKGVDPAGAIQRYKLTAATHALKKLLFPWGGG-------------
*: :**:.:: : ***. * : ::*.
Mflv_4847|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1586|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1669|M.smegmatis_MC2_155 --------------------------------------------------
TH_2183|M.thermoresistible__bu --------------------------------------------------
MAB_3676|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00696|M.leprae_Br4923 --------------------------------------------------
MAV_4300|M.avium_104 --------------------------------------------------
MMAR_1200|M.marinum_M --------------------------------------------------
MUL_1370|M.ulcerans_Agy99 --------------------------------------------------
Mb1579|M.bovis_AF2122/97 WARRRSYLRFMLREISCIFVAWFVLYLVLVLRAVGAGGNSYQRFLDFSAN
Rv1553|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4847|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1586|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1669|M.smegmatis_MC2_155 --------------------------------------------------
TH_2183|M.thermoresistible__bu --------------------------------------------------
MAB_3676|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00696|M.leprae_Br4923 --------------------------------------------------
MAV_4300|M.avium_104 --------------------------------------------------
MMAR_1200|M.marinum_M --------------------------------------------------
MUL_1370|M.ulcerans_Agy99 --------------------------------------------------
Mb1579|M.bovis_AF2122/97 PVVVVLNVVALSFLLLHAVTWFGSAPRAMVIQVRGRRVPARAVLAGHYAA
Rv1553|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4847|M.gilvum_PYR-GCK --------------
Mvan_1586|M.vanbaalenii_PYR-1 --------------
MSMEG_1669|M.smegmatis_MC2_155 --------------
TH_2183|M.thermoresistible__bu --------------
MAB_3676|M.abscessus_ATCC_1997 --------------
MLBr_00696|M.leprae_Br4923 --------------
MAV_4300|M.avium_104 --------------
MMAR_1200|M.marinum_M --------------
MUL_1370|M.ulcerans_Agy99 --------------
Mb1579|M.bovis_AF2122/97 WLVVSVIVAWMVLS
Rv1553|M.tuberculosis_H37Rv --------------