For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEPTVARPDIDPVLKMLLDTFPVTFTAADGVEVARARLRQLKTPPELLPELRIEERTVGYDGLTDIPVR VYWPPVVRDNLPVVVYYHGGGWSLGGLDTHDPVARAHAVGAQAIVVSVDYRLAPEHPYPAGIDDSWAALR WVGENAAELGGDPSRIAVAGDSAGGNISAVMAQLARDVGGPPLVFQLLWYPTTMADLSLPSFTENADAPI LDRDVIDAFLAWYVPGLDISDHTMLPTTLAPGNADLSGLPPAFIGTAEHDPLRDDGACYAELLTAAGVSV ELSNEPTMVHGYVNFALVVPAAAEATGRGLAALKRALHA
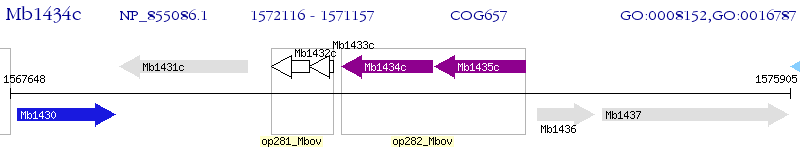
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1434c | lipH | - | 100% (319) | lipase LipH |
| M. bovis AF2122 / 97 | Mb1435c | lipI | 2e-97 | 58.10% (315) | lipase LipH |
| M. bovis AF2122 / 97 | Mb2994c | lipN | 1e-48 | 40.13% (319) | lipase/esterase LipN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3735 | - | 3e-94 | 57.47% (308) | alpha/beta hydrolase domain-containing protein |
| M. tuberculosis H37Rv | Rv1399c | lipH | 0.0 | 100.00% (319) | lipase LipH |
| M. leprae Br4923 | MLBr_00314 | - | 1e-15 | 28.57% (224) | putative esterase |
| M. abscessus ATCC 19977 | MAB_2814 | - | 1e-87 | 54.14% (314) | lipase LipH |
| M. marinum M | MMAR_2208 | lipH | 1e-144 | 77.02% (322) | lipase LipH |
| M. avium 104 | MAV_3377 | - | 2e-99 | 57.94% (321) | alpha/beta hydrolase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3059 | - | 6e-93 | 58.33% (300) | esterase |
| M. thermoresistible (build 8) | TH_3304 | - | 7e-61 | 62.69% (193) | PUTATIVE esterase |
| M. ulcerans Agy99 | MUL_1794 | lipH | 1e-143 | 76.71% (322) | lipase LipH |
| M. vanbaalenii PYR-1 | Mvan_2672 | - | 4e-93 | 56.37% (314) | alpha/beta hydrolase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1434c|M.bovis_AF2122/97 -------MTEPTVARPDIDPVLKMLLDTFPVTFTAADGVEVARARLRQLK
Rv1399c|M.tuberculosis_H37Rv -------MTEPTVARPDIDPVLKMLLDTFPVTFTAADGVEVARARLRQLK
MMAR_2208|M.marinum_M -------MTEQISTRPGIDPILKMLLDAVPMTFCAADGVELARERLAALK
MUL_1794|M.ulcerans_Agy99 -------MTEQISTRPGIDPILKMLLDAVPMTFCAADGVELARERLAALK
Mflv_3735|M.gilvum_PYR-GCK ------------MGMAALDPVLQTVLDAVPFQLTTDGGPEAARRRFRDLP
Mvan_2672|M.vanbaalenii_PYR-1 ----------MTVNPATLDAVLQKVLEAVPFKLTTEGGPEPARQRFKELP
MSMEG_3059|M.smegmatis_MC2_155 ----------------------------MPFQLSTADGVEEARRKFSELP
TH_3304|M.thermoresistible__bu --------------------------------------------------
MAV_3377|M.avium_104 ----MPRLDNPADEKPGIDPILQKVLDAVPFRLSTEDGIDAARQRFRDLP
MAB_2814|M.abscessus_ATCC_1997 -----------MTATAGIDPIIVKILEAVPIQLVSPDGPQASRDAYRAIA
MLBr_00314|M.leprae_Br4923 MAISAGAPVEIVESGPSIAARLASLTSRLTIRPILAVGSYVPQLPWPFGL
Mb1434c|M.bovis_AF2122/97 TPPELLPELR-IEERTVGYDGLTDIPVRVYWP-PVVRD-NLPVVVYYHGG
Rv1399c|M.tuberculosis_H37Rv TPPELLPELR-IEERTVGYDGLTDIPVRVYWP-PVVRD-NLPVVVYYHGG
MMAR_2208|M.marinum_M APPELLPDLR-TEDRKIGYGELTDIPVRIYWP-TVEPDRVLPVVVYYHGG
MUL_1794|M.ulcerans_Agy99 APPELLPDLR-TEDRKIGYGELTDIPVRTYWP-TVEPDWVLPVVVYYHGG
Mflv_3735|M.gilvum_PYR-GCK RS-PVHPDVE-TQDRAID-GPAGPIALRIYRPPTEQ--SVLPVVVFLHGG
Mvan_2672|M.vanbaalenii_PYR-1 RN-AILPEVA-AQDLSIE-GPAGPVPVRIYRPPSDE--AAPPTVVFIHGG
MSMEG_3059|M.smegmatis_MC2_155 RA-EIHPELS-VHDRTIE-GPAGPIGVRVYRPPTAEG-VKLPVVLFFHGG
TH_3304|M.thermoresistible__bu --------------------------------------------------
MAV_3377|M.avium_104 RR-PLHPELR-VEDRTIP-GPQGAIAVRIYWPPIHSESRPAPVVLYFHGG
MAB_2814|M.abscessus_ATCC_1997 TARPPWIEVANIEDRTIP-GPAGEIPVRIYSP---AEGGVRPVFVLIHGG
MLBr_00314|M.leprae_Br4923 IDHAARVLLPVTNAARAEVNLSNTSAQLVRAAGVRPADGSGRIVLYLHGG
Mb1434c|M.bovis_AF2122/97 GWSLGGLDTHDPVARAHAVGAQAIVVSVDYRLAPEHPYPAGIDDSWAALR
Rv1399c|M.tuberculosis_H37Rv GWSLGGLDTHDPVARAHAVGAQAIVVSVDYRLAPEHPYPAGIDDSWAALR
MMAR_2208|M.marinum_M GWALGSLDTHDHVARAHAVGAEAIVVSVDYRLAPEHPYPAGIEDSWAALR
MUL_1794|M.ulcerans_Agy99 GWALGSLDTHDHVARAHAVGAEAIVVSVDYRLAPEHPYPAGIEDSWAALR
Mflv_3735|M.gilvum_PYR-GCK GWTVGDLDTYDGQARMHAAGAGAVVVSVDYRLAPEHPYPAAVEDVWAATR
Mvan_2672|M.vanbaalenii_PYR-1 GWTVGDLDSYDGTARLHAAGAGAVVVSVDYRLAPEHPYPAAVDDVWAATR
MSMEG_3059|M.smegmatis_MC2_155 GWSVGDLDSYDATARRHAAGAEAVVVSVDYRLAPEHPYPAAVDDVWAATQ
TH_3304|M.thermoresistible__bu -----------------------------------------VEDCWAAVR
MAV_3377|M.avium_104 GFVIGDLDTHDGTARQHAVGAGAIVVSVDYRLAPEHPYPAAVEDAWAATL
MAB_2814|M.abscessus_ATCC_1997 GWVIGDLDTHDAVARAAAVQADAVVVSVDYRLAPEHPYPAAVEDCWAALQ
MLBr_00314|M.leprae_Br4923 AFLGCGANSHRRLVETLSKLADSPILVVNYRLLPKHSIGMALDDCHDGYQ
::* .
Mb1434c|M.bovis_AF2122/97 WVGENAAELGGDPSRIAVAGDSAGGNISAVMAQLARDVGG-----PPLVF
Rv1399c|M.tuberculosis_H37Rv WVGENAAELGGDPSRIAVAGDSAGGNISAVMAQLARDVGG-----PPLVF
MMAR_2208|M.marinum_M WVGEHAHELGGDPNRIAVAGDSAGGNISAIMAQLARDHAGN--GAPNLVF
MUL_1794|M.ulcerans_Agy99 WVGEHAHELGGDPNRIAVAGDSAGGNISAIMAQLARDHAGN--GAPNLVF
Mflv_3735|M.gilvum_PYR-GCK WVADNADQIGADADRVAVAGDSAGGNLAAVTAQSARDAG------IALRF
Mvan_2672|M.vanbaalenii_PYR-1 WVAENAAELGADPDRLAVAGDSAGGNLAAVVAQSARDAG------IPLRF
MSMEG_3059|M.smegmatis_MC2_155 WVAAHAEELGGDAERLAVAGDSAGGNLAAVVAQLARDAG-----APALRM
TH_3304|M.thermoresistible__bu WTAAHAAELGGDPARIAVAGDSAGGNLSAVMALLDRDHARADGSAPILRF
MAV_3377|M.avium_104 WAAENAAGLHGDPGRIAVAGDSAGGTLAAVTAQRARDRG-----GPPVRF
MAB_2814|M.abscessus_ATCC_1997 WVHEHAAEIGGDPGRLAVGGDSAGGNLSAVMAQLARDNG------IDLAF
MLBr_00314|M.leprae_Br4923 WLRR----LGYDPEQIVLAGDSAGGYLALALAQRLQDVGEE--PAALVAI
* : *. ::.:.****** :: * :* . : :
Mb1434c|M.bovis_AF2122/97 QLLWYPTTMADLSLPSFTENADAPILDRDVIDAFLAWYVPGLDISDHTML
Rv1399c|M.tuberculosis_H37Rv QLLWYPTTMADLSLPSFTENADAPILDRDVIDAFLAWYVPGLDISDHTML
MMAR_2208|M.marinum_M QLLWYPSCVGDLALPSFSENATAPILDLEVIDAFLSWYVPDLDVSDHTAL
MUL_1794|M.ulcerans_Agy99 QLLWYPSCVGDLALPSFSENATAPILDLEVIDAFLSWYVPDLDVSDHTAL
Mflv_3735|M.gilvum_PYR-GCK QLLWYPSTTFDTSLPSFRENADAPILDLASCKGFSRWYVGDLDLSGPSGP
Mvan_2672|M.vanbaalenii_PYR-1 QLLWYPSTTFDTSLPSFVENAQAPILDLAACKGFTRWYIGDTDLSEP---
MSMEG_3059|M.smegmatis_MC2_155 QLLWYPATTWDTSLPSFAENADAPVLAIDAVKGFSAWYAGHVDLTDP---
TH_3304|M.thermoresistible__bu QLLWYPATTWDTSLPSFSENADAPVLGLDAVAGLSRWYAGHVDLSDP---
MAV_3377|M.avium_104 QLLWYPSTMWDASLPSFAENATAPILDVKAVAEFSRWYAGDVDLSDP---
MAB_2814|M.abscessus_ATCC_1997 QVLWYPATTFDTTLPSMVENAEAPVLDFKTVGALSKWYLGDTNPATAG--
MLBr_00314|M.leprae_Br4923 SPLLQLAKKGKQAHPNAKTDAMFPPKAFDALGRLVASSAAKNKVDDK---
. * : . : *. :* * : .
Mb1434c|M.bovis_AF2122/97 PTTLAPGNA-DLSGLPPAFIGTAEHDPLRDDGACYAELLTAAGVSVELSN
Rv1399c|M.tuberculosis_H37Rv PTTLAPGNA-DLSGLPPAFIGTAEHDPLRDDGACYAELLTAAGVSVELSN
MMAR_2208|M.marinum_M PATLAPGNG-DLTGLPPAFIGTAEHDPLRDDGARYAELLTAAGIAAEWCN
MUL_1794|M.ulcerans_Agy99 PATLAPGNG-DLTGLPPAFIGTAEHDPLRDDGARYAELLTAAGIAAEWCN
Mflv_3735|M.gilvum_PYR-GCK PVTLVPARG-DLTGLPPAYIAVAGHDPLRDDGVRYAELLTAAGVPVELHH
Mvan_2672|M.vanbaalenii_PYR-1 PVTLVPARG-NLAGLPPAYIAVAGHDPLRDDGLRYGELLTAAGVPVELHH
MSMEG_3059|M.smegmatis_MC2_155 PATLVPARHDDLSGLAPAYIAVAGHDPLRDDGVHYGDLLAAAGVPVQLDN
TH_3304|M.thermoresistible__bu PVTLVPARADDLSGLPPAYIAVAGHDPLRDDGAKYAELLTAAGVPVELHN
MAV_3377|M.avium_104 PADMAPGRATDLSNLPPAYIGVAGYDPLRDDGIRYGERLAAAGVAAQVHN
MAB_2814|M.abscessus_ATCC_1997 -PTLVPARAENFEGLAPAFIGTAQFDPLRDDGAKYAELLRDAGVPVQLHN
MLBr_00314|M.leprae_Br4923 PEELYEPLDHIAPGLPRTLIHVSGSEVLLHDAQLAATKLAAVGVPAEIRV
: .*. : * .: : * .*. . * .*:..:
Mb1434c|M.bovis_AF2122/97 EPTMVHGYVNFALVVPAAAEATGRGLAALKRALHA---------
Rv1399c|M.tuberculosis_H37Rv EPTMVHGYVNFALVVPAAAEATGRGLAALKRALHA---------
MMAR_2208|M.marinum_M EPNLVHGYVSFAVVVPSAAEATSRGLAALRKVLHY---------
MUL_1794|M.ulcerans_Agy99 EPNLVHGYVSFAVVVPSAAEATSRGLATLRKVLHY---------
Mflv_3735|M.gilvum_PYR-GCK AESLVHGYLGYAGIVPAATEATERGLEALRAALA----------
Mvan_2672|M.vanbaalenii_PYR-1 AESLVHGYLGYAGIVPAATEATERGLAALRAALA----------
MSMEG_3059|M.smegmatis_MC2_155 ADTLVHGYLGYSGVVPAATEAFDRGLAALREALHS---------
TH_3304|M.thermoresistible__bu ADTLVHGYLGYSGVVPAATEATERALKALRAALHGPGSAS----
MAV_3377|M.avium_104 AETLVHGYLGYAGVVPAATAAMERGLVALRTALQG---------
MAB_2814|M.abscessus_ATCC_1997 APTLIHGYLGYADIVPSATEERDLSLAALKQALHG---------
MLBr_00314|M.leprae_Br4923 WPGQVHVFQAAAPMVPEATLSLRQISQYIREATASKASRDRQSL
:* : : :** *: :: .