For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVRADRDRWDLATSVGATATMVAAQRALAADPRYALIDDPYAAPLVRAVGMDVYTRLVDWQIPVEGDSEF DPQRMATGMACRTRFFDQFFLDATHSGIGQFVILASGLDARAYRLAWPVGSIVYEVDMPEVIEFKTATLS DLGAEPATERRTVAVDLRDDWATALQTAGFDPKVPAAWSAEGLLVYLPVEAQDALFDNITALSAPGSRLA FEFVPDTAIFADERWRNYHNRMSELGFDIDLNELVYHGQRGHVLDYLTRDGWQTSALTVTQLYEANGFAY PDDELATAFADLTYSSATLMR
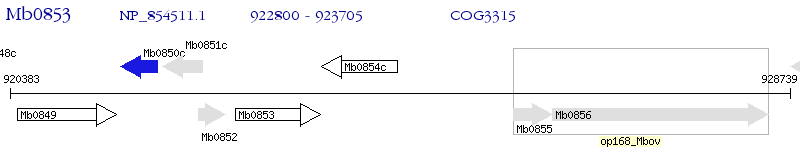
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0853 | - | - | 100% (301) | hypothetical protein Mb0853 |
| M. bovis AF2122 / 97 | Mb1758c | - | 2e-82 | 53.93% (280) | hypothetical protein Mb1758c |
| M. bovis AF2122 / 97 | Mb3432 | - | 1e-81 | 53.27% (306) | hypothetical protein Mb3432 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5025 | - | 6e-82 | 53.47% (303) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv0830 | - | 1e-175 | 100.00% (301) | hypothetical protein Rv0830 |
| M. leprae Br4923 | MLBr_02640 | - | 5e-60 | 46.59% (279) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_3787 | - | 1e-77 | 50.32% (312) | hypothetical protein MAB_3787 |
| M. marinum M | MMAR_4850 | - | 1e-139 | 78.74% (301) | O-methyltransferase |
| M. avium 104 | MAV_0778 | - | 1e-127 | 72.58% (299) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_1479 | - | 4e-87 | 53.64% (302) | methyltransferase, putative, family protein |
| M. thermoresistible (build 8) | TH_2706 | - | 2e-81 | 54.08% (294) | PUTATIVE putative methyltransferase |
| M. ulcerans Agy99 | MUL_0450 | - | 1e-135 | 76.08% (301) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1344 | - | 2e-80 | 52.16% (301) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0853|M.bovis_AF2122/97 MVRADRDRWDLATSVGATATMVAAQRALAADPRYALIDDPYAAPLVRAVG
Rv0830|M.tuberculosis_H37Rv MVRADRDRWDLATSVGATATMVAAQRALAADPRYALIDDPYAAPLVRAVG
MMAR_4850|M.marinum_M MVRTDRDRWDLATSVGATATMVAAQRALAADPQYALIDDPYAAPLVRAVG
MUL_0450|M.ulcerans_Agy99 MVRTDRDRWDLATSVGATATMVAAQRALAADPQYALIDDPYAAPLVRAVD
MAV_0778|M.avium_104 MARTDHDRWDLATSVGATATMVAAQRALSSDAN--LIDDPYAAPLVRAVG
TH_2706|M.thermoresistible__bu MARSESDTWDLTSGVGVTATMVAAARALASREPEPLIDDPFAAPLVRAVG
Mflv_5025|M.gilvum_PYR-GCK MARTPDDSWDLASSVGATATMVAAGRAVASADPDPLINDPYAEPLVRAVG
Mvan_1344|M.vanbaalenii_PYR-1 MARTADDSWDPASSVGATATMVAAGRAAASADPDPLINDPYAEPLVRAVG
MSMEG_1479|M.smegmatis_MC2_155 MVRSDNDTWDLASSVGATATMVAAGRAVVSQDPGGLINDPFAAPLVRAVG
MAB_3787|M.abscessus_ATCC_1997 MARTDDDSWDLASSVGATATMVAAQRALASHVPNPLINDPYAEPLVRAVG
MLBr_02640|M.leprae_Br4923 -MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTNTG
*: * ** :.**.**.**** ** : ** **:* ** ..
Mb0853|M.bovis_AF2122/97 M-DVYTRLVDWQIPVEGDSEFD-----PQRMATGMACRTRFFDQFFLDAT
Rv0830|M.tuberculosis_H37Rv M-DVYTRLVDWQIPVEGDSEFD-----PQRMATGMACRTRFFDQFFLDAT
MMAR_4850|M.marinum_M I-DVYTRLVNGQIPVDVESGFD-----PARMAEAMACRTRFYDQFFVEAT
MUL_0450|M.ulcerans_Agy99 I-DVYTRLVNGQIPVDVESGFD-----PARMPEAMACRTRFYDQFFVDAT
MAV_0778|M.avium_104 I-DVYVRLVDGEIQP-GTSEFD-----PHRMAKGMACRTRFYDDFFLDAA
TH_2706|M.thermoresistible__bu V-EFFTRLVDGDLELPADTAEERA--AAQVLTEVMAVRTRFFDDFFTDAT
Mflv_5025|M.gilvum_PYR-GCK L-DFFTRMLDGELDLSVFPDSSPE--RAQAMIDGMAVRTRFFDDCCLAAA
Mvan_1344|M.vanbaalenii_PYR-1 L-SFFTKMLDGELDLSQFADGSPE--RVQAMIDGMAVRTRFFDDCCGTST
MSMEG_1479|M.smegmatis_MC2_155 I-EALTMLADGKFDIEKVLPESAA--RVRANIDEMAVRTKFFDDYFMDAT
MAB_3787|M.abscessus_ATCC_1997 I-EYFTKLASGAFDPEQMAGLVQLGITADGMANGMASRTWYYDTAFESST
MLBr_02640|M.leprae_Br4923 AGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNNAV
: . : * ** ::* :.
Mb0853|M.bovis_AF2122/97 HSGIGQFVILASGLDARAYRLAWPVGSIVYEVDMPEVIEFKTATLSDLGA
Rv0830|M.tuberculosis_H37Rv HSGIGQFVILASGLDARAYRLAWPVGSIVYEVDMPEVIEFKTATLSDLGA
MMAR_4850|M.marinum_M RSGISQVVILASGLDARAYRLDWPAGTVVYEVDMPEVIEFKTLTLADLGA
MUL_0450|M.ulcerans_Agy99 RSGISQVVILASGLDARAYRLGWPAGTVVHEVDMPQVIEFKTLTLADLGA
MAV_0778|M.avium_104 RAGVGQAVILASGLDARAYRLPWPAGTVVYEVDMPDVIEFKTLTLADLGA
TH_2706|M.thermoresistible__bu RVGARQAVILASGLDARAYRLTWPDGTVVYEIDQPEVIAFKTRTLAELGA
Mflv_5025|M.gilvum_PYR-GCK SAGVRQVVILAAGLDARTYRLPWPDGTVVYELDQPDVIAFKTQTLRNLGA
Mvan_1344|M.vanbaalenii_PYR-1 TAGITQVVILASGLDARAYRLGWPAGTVVYELDQPAVIEFKTRTLAGLGA
MSMEG_1479|M.smegmatis_MC2_155 GRGVGQAVILASGLDSRAYRLPWPDGTVVYEIDQPDVIEFKTRTLADLGA
MAB_3787|M.abscessus_ATCC_1997 AAGVRQAVILASGLDTRAYRLTWPAGTTVYELDQPEVITFKTDTLAELGA
MLBr_02640|M.leprae_Br4923 IDGIRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTLAEHGV
* * ****:***:*:*** ** *: *:*:* * *: :*: ** *.
Mb0853|M.bovis_AF2122/97 EPATERRTVAVDLRDDWATALQTAGFDPKVPAAWSAEGLLVYLPVEAQDA
Rv0830|M.tuberculosis_H37Rv EPATERRTVAVDLRDDWATALQTAGFDPKVPAAWSAEGLLVYLPVEAQDA
MMAR_4850|M.marinum_M EPTAERRTVAVDLRDDWAAALQAAGFDKDVPSAWSAEGLLVYLPDDAQDA
MUL_0450|M.ulcerans_Agy99 KPTAERRTVAVDLRDDWAAVLQAAGFDKDVPSAWSAEGLLVYLPDDAQGA
MAV_0778|M.avium_104 QPTAQRRTVAIDLRDDWAAALREEGFDTQAPAAWSAEGLLVYLPEQAQDA
TH_2706|M.thermoresistible__bu PPTAPHRPVAVDLRDDWPAALTAAGFDPTVATAWSAEGLLRYLPPDAQDR
Mflv_5025|M.gilvum_PYR-GCK EPAATQRPVPVDLREDWPAALRAAGFDATRPTAWLAEGLLIYLPPEAQDA
Mvan_1344|M.vanbaalenii_PYR-1 HPTATHRPVPIDLREDWPAALHAAGFDASRPTAWLAEGLLIYLPPEAQDQ
MSMEG_1479|M.smegmatis_MC2_155 EPTCERRTVSIDLRDDWPAALRAAGFDPSAPTAWCAEGLLIYLPPEAQDK
MAB_3787|M.abscessus_ATCC_1997 SPSAERRTVAIDLREDWPAALQAAGFDPEQPTVWSAEGLLIYLPPEAQDR
MLBr_02640|M.leprae_Br4923 TPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPATAQDG
*: :* *.:***:**...* *** :.* ***** *** **.
Mb0853|M.bovis_AF2122/97 LFDNITALSAPGSRLAFEFVPDTAIFADERWRNYHNRMS-ELGFD--IDL
Rv0830|M.tuberculosis_H37Rv LFDNITALSAPGSRLAFEFVPDTAIFADERWRNYHNRMS-ELGFD--IDL
MMAR_4850|M.marinum_M LFDNITALSATGSRLAFEFVPDTAVFNDERWRSHHARMS-ELGFE--IDF
MUL_0450|M.ulcerans_Agy99 LFDNVTALSATGSRLAFEFVPDTAVFNDERWRSHHARMS-ELGFE--IDF
MAV_0778|M.avium_104 LFDNITALSAPGSRLAFDFVPDTAVFADPRWRAHHERMS-ELGFE--VDF
TH_2706|M.thermoresistible__bu LFDHITDLSAPGSRLATEDHPGPGAGLGEKADELGRRWR-EHGFE--VQL
Mflv_5025|M.gilvum_PYR-GCK LFDTVTALSAPGSTLATEYVPGIVDFDGAKARALSEPLR-EHGLD--IDM
Mvan_1344|M.vanbaalenii_PYR-1 LFDRITALSAPGSTIATEYAPGIIDFDDEKARAMSAPMR-EMGLD--IDM
MSMEG_1479|M.smegmatis_MC2_155 LFDDIHHLSAPGSTVATEFVPALKDFDPEKARQATETLT-RMGVD--IDM
MAB_3787|M.abscessus_ATCC_1997 LFASITELSAPGSRLVCEQVPGLETADFSKARELTRQFA-GETLD--LDL
MLBr_02640|M.leprae_Br4923 LFTEIGGLSAVGSRIAVETSPLHGDEWREQMQLRFRRVSDALGFEQAVDV
** : *** ** :. : * : .: ::.
Mb0853|M.bovis_AF2122/97 NELVYHGQ-RGHVLDYLTRDGWQTSALTVTQLYEANGFAYP-DDELATAF
Rv0830|M.tuberculosis_H37Rv NELVYHGQ-RGHVLDYLTRDGWQTSALTVTQLYEANGFAYP-DDELATAF
MMAR_4850|M.marinum_M NDLVYHGQ-RSHVIDHLARDGWQSASHTAKELHAANGFDYP-DDDIAAVF
MUL_0450|M.ulcerans_Agy99 NDLVYHGQ-RSHVIDHLARDGWQSASHTAKELHAANGLDYP-DDDIAAVF
MAV_0778|M.avium_104 NDLVYHGE-RSHIVDHLSGRGWRVTSRTIAELHAANGFAYA-ADDVAAAF
TH_2706|M.thermoresistible__bu SELFHPGE-RTPVVDYLRGRGWDARARPRAEVFAGYGREFP-DSAALAAM
Mflv_5025|M.gilvum_PYR-GCK PSLVYTGA-RTPVMDHLRGAGWQVSGTARAELFARFGRPLPQDADGTDPL
Mvan_1344|M.vanbaalenii_PYR-1 PSLVYAGT-RSHVMDYLAGQGWEVTGVPRRELFTRYGRPLPPEAGDTDPL
MSMEG_1479|M.smegmatis_MC2_155 PSLIYHGE-RHSASDYLETKGWQMDSAARATLFTRYGLPAPGHDDD-DPL
MAB_3787|M.abscessus_ATCC_1997 ESLVYTQE-RQMAADWLAEHGWTVITEENDELYARLNLDPPNPLLR-SIF
MLBr_02640|M.leprae_Br4923 QELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGRWGDGVPMADDKD
.*.: * * * ** .
Mb0853|M.bovis_AF2122/97 ADLTYSSATLMR------
Rv0830|M.tuberculosis_H37Rv ADLTYSSATLMR------
MMAR_4850|M.marinum_M ADITYTSAVLGR------
MUL_0450|M.ulcerans_Agy99 ADITYTSAVLGR------
MAV_0778|M.avium_104 ADVTYSSAVLGG------
TH_2706|M.thermoresistible__bu RDSSAVTAVRPVVRAGMR
Mflv_5025|M.gilvum_PYR-GCK GEIVYVSATLAADA----
Mvan_1344|M.vanbaalenii_PYR-1 GEIVYVSAHRPQ------
MSMEG_1479|M.smegmatis_MC2_155 GEIIYISGTLR-------
MAB_3787|M.abscessus_ATCC_1997 PNIVYVDATLG-------
MLBr_02640|M.leprae_Br4923 AFAEFVTAHRL-------
.