For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STESTSPAIVVGVDGSPASTLAVDWAARDAEMRNVPLKVVHVFAPMVAPANGWADVAMPADYARWREDQA HQIVQQAHAVAVDAAAPGRAAHVTSEVRYAAIVPAMVELSLQAEMVVVGCRGQGGVASALLGSVSSGLAH HAHCPVAIIHDDEPLPPRSAQAPVLVGVDGSPTSELATEIGFDEASRRGAELVALHAWTDMGPLDFPSIN WAPIEWSNIKDQEEEVLAERLCGWQERYPDVVTHKVVVSDRPAPRLLEHAHDAQLVVVGSHGRGGFPGML LGSVSRAVVNSARIPVIIARTPANS*
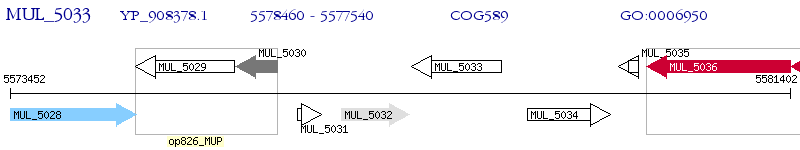
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_5033 | - | - | 100% (306) | hypothetical protein MUL_5033 |
| M. ulcerans Agy99 | MUL_2234 | - | 8e-86 | 52.98% (302) | hypothetical protein MUL_2234 |
| M. ulcerans Agy99 | MUL_3267 | - | 2e-81 | 50.17% (301) | hypothetical protein MUL_3267 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2019 | - | 1e-109 | 63.29% (316) | hypothetical protein Mb2019 |
| M. gilvum PYR-GCK | Mflv_4382 | - | 6e-83 | 52.65% (302) | UspA domain-containing protein |
| M. tuberculosis H37Rv | Rv1996 | - | 1e-109 | 63.29% (316) | hypothetical protein Rv1996 |
| M. leprae Br4923 | MLBr_01390 | - | 3e-06 | 29.17% (144) | hypothetical protein MLBr_01390 |
| M. abscessus ATCC 19977 | MAB_2489 | - | 5e-47 | 36.63% (303) | hypothetical protein MAB_2489 |
| M. marinum M | MMAR_5410 | - | 1e-173 | 97.71% (306) | hypothetical protein MMAR_5410 |
| M. avium 104 | MAV_2507 | - | 2e-87 | 53.82% (301) | universal stress protein family protein |
| M. smegmatis MC2 155 | MSMEG_3940 | - | 2e-73 | 49.66% (294) | universal stress protein family protein |
| M. thermoresistible (build 8) | TH_2494 | - | 6e-76 | 49.53% (317) | PUTATIVE putative conserved hypothetical membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1963 | - | 1e-82 | 54.82% (301) | UspA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4382|M.gilvum_PYR-GCK --MSTQPAPLGIVVGVDGSAASRVAVDWAARDAALRQVPLTLAYVLPGAA
Mvan_1963|M.vanbaalenii_PYR-1 --MSTQPAPVGIVVAVDGSDASRVAVDWAARDAAMRRIPLTLAHVLPSAA
MUL_5033|M.ulcerans_Agy99 --MSTESTSPAIVVGVDGSPASTLAVDWAARDAEMRNVPLKVVHVFAPMV
MMAR_5410|M.marinum_M --MSTESTSPAIVVGVDGSPASTLAVEWAARDAEMRNVPLKVVHVFAPMV
Mb2019|M.bovis_AF2122/97 --MSAQQTNLGIVVGVDGSPCSHTAVEWAARDAQMRNVALRVVQVVPPVI
Rv1996|M.tuberculosis_H37Rv --MSAQQTNLGIVVGVDGSPCSHTAVEWAARDAQMRNVALRVVQVVPPVI
MSMEG_3940|M.smegmatis_MC2_155 MSHNGNGAGYGMVVAVDGSAESDAAVRWAAREATLRKIPVTVMHVVEPMI
TH_2494|M.thermoresistible__bu --VSSTAAYEGIVVGVDGSAASADAVRWAARSAALRNLQLTLVHVMTATS
MAV_2507|M.avium_104 --MAASSKRCGVLVGVDGSPASNFAVCWAARDAAMRNVPLTLVHMVNAAT
MAB_2489|M.abscessus_ATCC_1997 --MTTSTPNSHILVGVDGSTSALHALTWAGAEAQRRNLPLTLVHVIDHSS
MLBr_01390|M.leprae_Br4923 -----MGAYQTVVVGTDGSDSSLRAVDRAGKIAG----------------
::*..*** : *: *. *
Mflv_4382|M.gilvum_PYR-GCK VQS---WIQVPLPASFYEDEK--------AEATRVLADARAVVDAATQAH
Mvan_1963|M.vanbaalenii_PYR-1 TQS---WIQVPLPAAFFEDEK--------LEAERILADARALVTAATEGG
MUL_5033|M.ulcerans_Agy99 APANG-WADVAMPADYARWRE--------DQAHQIVQQAHAVAVDAAAPG
MMAR_5410|M.marinum_M TPANG-WADVAMPADYARWRE--------DQAHQIVQQAHAVAVDAAAPG
Mb2019|M.bovis_AF2122/97 TAPEG-WAFEYSRFQEAQKREIVEHSYLVAQAHQIVEQAHKVALEASSSG
Rv1996|M.tuberculosis_H37Rv TAPEG-WAFEYSRFQEAQKREIVEHSYLVAQAHQIVEQAHKVALEASSSG
MSMEG_3940|M.smegmatis_MC2_155 VN----WPVPPVQGSVTEWQE--------ANARNVIKH----AHDTFVAV
TH_2494|M.thermoresistible__bu IGAPLVWPAGPIPDSLIQWQE--------DEARQILADSAEIAENAFQEA
MAV_2507|M.avium_104 V-----WPQVPMAAEAVAWQE--------DDGRRVLQEAVKIAEDATRNG
MAB_2489|M.abscessus_ATCC_1997 LGQG---YNLGASASFFQHLE--------TDGAEFLSQAKDHVYALHPNV
MLBr_01390|M.leprae_Br4923 -----------------------------SDAKLIIASAYLPQHDN----
:. .:
Mflv_4382|M.gilvum_PYR-GCK PLPAVT----------EKVLSGQPVPTLADLSRDADMIVVGSRGMGKWER
Mvan_1963|M.vanbaalenii_PYR-1 EALTVD----------EVVLSGQPVAALVDLAKDAEMLVMGSRGLGKWER
MUL_5033|M.ulcerans_Agy99 RAAHVT----------SEVRYAAIVPAMVELSLQAEMVVVGCRGQGGVAS
MMAR_5410|M.marinum_M RAAHVT----------SEVRYAAIVPAMVELSQQAEMVVVGCRGQGGVAS
Mb2019|M.bovis_AF2122/97 RAAQIT----------GEVLHGQIVPTLANISRQVAMVVLGYRGQGAVAG
Rv1996|M.tuberculosis_H37Rv RAAQIT----------GEVLHGQIVPTLANISRQVAMVVLGYRGQGAVAG
MSMEG_3940|M.smegmatis_MC2_155 EESAP-----DGIR--HEIRYAGIVAELVDVSKNATMMVVGSRGLGAFGG
TH_2494|M.thermoresistible__bu RAAGPGPAGDDAVRVVTGVVRAAPVPTLVDASKQAKLVAVGSRGQGPLRR
MAV_2507|M.avium_104 RNLAIT----------TELWHAPPAPTLAQLSEEAELVVVGSTGRGAIGR
MAB_2489|M.abscessus_ATCC_1997 EVSTVK-------------ATGRPVPVLVELSRNALMTVLGSSGLGGFTG
MLBr_01390|M.leprae_Br4923 -------------------------ARAFDILKDESYKVTG---------
. : : . *
Mflv_4382|M.gilvum_PYR-GCK RVLGSVSSGLVHHARCPVAVIYDEDPLIPHPAQAPVVVGIDGSPASERAT
Mvan_1963|M.vanbaalenii_PYR-1 RLLGSVSSGVVHHAHCPVAVIHDVDPLIPHPAQAPVVVGVDGSPASEHAT
MUL_5033|M.ulcerans_Agy99 ALLGSVSSGLAHHAHCPVAIIHDDEPLPPRSAQAPVLVGVDGSPTSELAT
MMAR_5410|M.marinum_M ALLGSVSSGLAHHAHCPVAIIHDDEALPPRSAQAPVLVGVDGSPTSELAT
Mb2019|M.bovis_AF2122/97 ALLGSVSSSLVRHAHGPVAVIPE-EPRPARPPHAPVVVGIDGSPTSGLAA
Rv1996|M.tuberculosis_H37Rv ALLGSVSSSLVRHAHGPVAVIPE-EPRPARPPHAPVVVGIDGSPTSGLAA
MSMEG_3940|M.smegmatis_MC2_155 ALLGSVSSGVIHHAHCPVAVIHGDQIRRPD-PRSPVLVGIDGSPASEDAT
TH_2494|M.thermoresistible__bu VLLGSVSSGLVHHAHCPVAVIREGTPHTPDFAKRPVLVGIDGSESAQRAT
MAV_2507|M.avium_104 LLLGSVSSGLVRRARCPVAVIHDEDPMMPYPQRAPVLVGIDGSPASELAT
MAB_2489|M.abscessus_ATCC_1997 MMAGSVSVSLTAKGHSPVVVVRE----SPVPGGGPVVVGIDDSESSDGAA
MLBr_01390|M.leprae_Br4923 --------------TAPIYEILH-----------------DAKERAHAAG
*: : * . : *
Mflv_4382|M.gilvum_PYR-GCK AIAFDEACRRGVDLVAVHTWSDAG--YELP-----GVEWTEVQPEEEMLL
Mvan_1963|M.vanbaalenii_PYR-1 AIAFDEASFRGVDLVAVHTWSDAG--YELP-----GTGWAEIQPEEDMLL
MUL_5033|M.ulcerans_Agy99 EIGFDEASRRGAELVALHAWTDMGP-LDFPSINWAPIEWSNIKDQEEEVL
MMAR_5410|M.marinum_M EIAFDEASRRGAELVALHAWTDMGP-LDFPSINWAPIEWSNIKDQEEEVL
Mb2019|M.bovis_AF2122/97 EIAFDEASRRGVDLVALHAWSDMGP-LDFPRLNWAPIEWRNLEDEQEKML
Rv1996|M.tuberculosis_H37Rv EIAFDEASRRGVDLVALHAWSDMGP-LDFPRLNWAPIEWRNLEDEQEKML
MSMEG_3940|M.smegmatis_MC2_155 AFAFDEASRRRVDLIALHAWSDVGV-FSAL-----GMDWHEYEDQGRELL
TH_2494|M.thermoresistible__bu AVAFEEASFRGVDLIALHAWSDADI-ADIP-----TLELSAVQQSAERVL
MAV_2507|M.avium_104 AIAFDEALRRGVDLNAVHAWSDTQV-FGLP-----GMDWPAVRSQAERSL
MAB_2489|M.abscessus_ATCC_1997 AWAFEEAAMRETELVAVHVWNNIPPGYVYAYTAWSAMDSAGEQQRQEQLL
MLBr_01390|M.leprae_Br4923 AKNVEERPVVGAPVDALVNLAEKTN-------------------------
.:* . : *: :
Mflv_4382|M.gilvum_PYR-GCK SERLAGWHERYPDVTVHRVVRRDQPARRLLEEAEKAQLLVVGSHGRGGFT
Mvan_1963|M.vanbaalenii_PYR-1 AERLAGWQERYPDVTVHRVVCRDQPARRLLEEAAKGQLLVVGSHGRGGFG
MUL_5033|M.ulcerans_Agy99 AERLCGWQERYPDVVTHKVVVSDRPAPRLLEHAHDAQLVVVGSHGRGGFP
MMAR_5410|M.marinum_M AERLCGWQERYPDVVTHKVVVSDRPAPRLLEHAHDAQLVVVGSHGRGGFP
Mb2019|M.bovis_AF2122/97 ARRLSGWQDRYPDVVVHKVVVCDRPAPRLLELAQTAQLVVVGSHGRGGFP
Rv1996|M.tuberculosis_H37Rv ARRLSGWQDRYPDVVVHKVVVCDRPAPRLLELAQTAQLVVVGSHGRGGFP
MSMEG_3940|M.smegmatis_MC2_155 GERLAGWRERYPDVRVTRQIVCDQPARWLIDGSRHAQLVVVGSHGRAGFA
TH_2494|M.thermoresistible__bu SERLAGFRERYPDVSVHRRIVGSEPARHLVEASEDAQLVVVGSRGRGGFT
MAV_2507|M.avium_104 AERLAGWQERYPDVTVHRMVVCDRPARQLIEQSESAQLTVVGSHGRGGLA
MAB_2489|M.abscessus_ATCC_1997 AERLAGWQEKYPDVPVRRVVAVGHPADVLLSQAAGAQLIVTGSRGRGDIA
MLBr_01390|M.leprae_Br4923 -----------------------------------ADLLVVGNVGLSTIA
.:* *.*. * . :
Mflv_4382|M.gilvum_PYR-GCK GMLLGSVGSQIVQSSRTPVIVARTA-------
Mvan_1963|M.vanbaalenii_PYR-1 GMLLGSVGSQVVQSARGPVIVARR--------
MUL_5033|M.ulcerans_Agy99 GMLLGSVSRAVVNSARIPVIIARTPANS----
MMAR_5410|M.marinum_M GMLLGSVSRAVINSARIPVIIARTPPNS----
Mb2019|M.bovis_AF2122/97 GMHLGSVSRAVVNSGQAPVIVARIPQDPAVPA
Rv1996|M.tuberculosis_H37Rv GMHLGSVSRAVVNSGQAPVIVARIPQDPAVPA
MSMEG_3940|M.smegmatis_MC2_155 GMLLGSVGSKVARAAYSPVIVVR---------
TH_2494|M.thermoresistible__bu GMLLGSVSRAVVNAVTTPVIVAR---------
MAV_2507|M.avium_104 GTLLGSVSNAVVHSVRMPVIVARPS-------
MAB_2489|M.abscessus_ATCC_1997 GFFLGSTSHALIHKASCPVLVAPRT-------
MLBr_01390|M.leprae_Br4923 GRLLGSVPANVSRRAKIDVLIVHTTH------
* ***. : . *::.