For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TASQLFRRAKVIEEVIDLIADVGADAVQMRDVAQRSGVALATVYRYFSSKEHLLAAALQDWQMRLTRRIL SASGSPDQDPLPGILDYLRRALRAFHRNPEMTALMLQMMTSTDRDAKATIDQMDRTNVEMFNRLLHGVAP ADISNVSFALNAALSGALTGLLTSRLTLEEALGHVEWVARALLDEGRPHP*
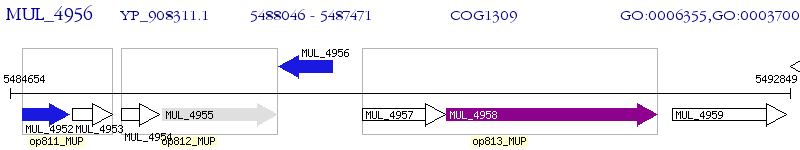
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4956 | - | - | 100% (191) | TetR family transcriptional regulator |
| M. ulcerans Agy99 | MUL_4145 | - | 1e-14 | 29.79% (188) | TetR family transcriptional regulator |
| M. ulcerans Agy99 | MUL_0681 | - | 3e-09 | 33.33% (96) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3605 | - | 1e-14 | 29.79% (188) | transcriptional regulatory protein TetR-family |
| M. gilvum PYR-GCK | Mflv_1459 | - | 3e-15 | 29.26% (188) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3574 | - | 1e-14 | 29.79% (188) | transcriptional regulatory protein TetR-family |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0579c | - | 2e-14 | 29.48% (173) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0141 | - | 1e-104 | 100.00% (191) | TetR family transcriptional regulator |
| M. avium 104 | MAV_0584 | - | 3e-15 | 30.85% (188) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6042 | - | 2e-14 | 30.11% (186) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_1745 | - | 6e-16 | 29.69% (192) | TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY) |
| M. vanbaalenii PYR-1 | Mvan_5312 | - | 2e-14 | 28.72% (188) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3605|M.bovis_AF2122/97 ---------------------------------------------MAVLA
Rv3574|M.tuberculosis_H37Rv ---------------------------------------------MAVLA
MAV_0584|M.avium_104 ---------------------------------------------MAVLA
Mflv_1459|M.gilvum_PYR-GCK ------------MSGSSESAT------------AGSDSRPRQVMNVAVLA
Mvan_5312|M.vanbaalenii_PYR-1 ------------MSGTSEFAADKSQTSARATSGAGSESRPRQVMNVAVLA
MSMEG_6042|M.smegmatis_MC2_155 ------------------------------------------MTNVAVLS
TH_1745|M.thermoresistible__bu MSSSTGTNPGRGSDSTGSETNGAAATGAGSGSTGTSRTRGRQVTNVAVLT
MAB_0579c|M.abscessus_ATCC_199 -----------------MATPRTSSASEAAQSGDGDTDATSQPAAVSALS
MUL_4956|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0141|M.marinum_M --------------------------------------------------
Mb3605|M.bovis_AF2122/97 ESELGSEAQRERRKRILDATMAIASKGGYEAVQMRAVADRADVAVGTLYR
Rv3574|M.tuberculosis_H37Rv ESELGSEAQRERRKRILDATMAIASKGGYEAVQMRAVADRADVAVGTLYR
MAV_0584|M.avium_104 ESELGSEAQRERRKRILDATMAIASKGGYEAVQMRAVADRADVAVGTLYR
Mflv_1459|M.gilvum_PYR-GCK ESELGSEAQRERRKRILDATLAIASKGGYEAVQMRAVAERADVAVGTLYR
Mvan_5312|M.vanbaalenii_PYR-1 ESELGSEAQRERRKRILDATLAIASKGGYEAVQMRAVAERADVAVGTLYR
MSMEG_6042|M.smegmatis_MC2_155 ESELGSEAQRERRKRILDATLAIASKGGYEAVQMRAVAERADVAVGTLYR
TH_1745|M.thermoresistible__bu DSELGSEAQRERRKRILDATLAIASKGGYEAVQMRAVAERADVAVGTLYR
MAB_0579c|M.abscessus_ATCC_199 DDDLGSSAQRERRKRILDATLAIASKGGYEAVQMRTVAERADVAVGTLYR
MUL_4956|M.ulcerans_Agy99 ----MTASQLFRRAKVIEEVIDLIADVGADAVQMRDVAQRSGVALATVYR
MMAR_0141|M.marinum_M ----MTASQLFRRAKVIEEVIDLIADVGADAVQMRDVAQRSGVALATVYR
: :* ** :::: .: : :. * :***** **:*:.**:.*:**
Mb3605|M.bovis_AF2122/97 YFPSKVHLLVSALGREFSRIDAKTDRSAVAGA-TPFQRLNFMVGKLNRAM
Rv3574|M.tuberculosis_H37Rv YFPSKVHLLVSALGREFSRIDAKTDRSAVAGA-TPFQRLNFMVGKLNRAM
MAV_0584|M.avium_104 YFPSKVHLLVSALGREFSRIDAKTDRAALTGG-TPFQRLNFMVGKLNRAM
Mflv_1459|M.gilvum_PYR-GCK YFPSKVHLLVSALGREFERIDAKTDRAALAAGATAYQRLNIMVGKLNRAM
Mvan_5312|M.vanbaalenii_PYR-1 YFPSKVHLLVSALGREFERIDAKTDRSAFPAGATAYQRLNIMVGKLNRAM
MSMEG_6042|M.smegmatis_MC2_155 YFPSKVHLLVSALGREFERIDAKTDRAALAGG-TPYQRLNFMVGKLNRAM
TH_1745|M.thermoresistible__bu YFPSKVHLLVSALGREFERIDAKTDRSTLVGD-TPYERLNFMVSRLNRAM
MAB_0579c|M.abscessus_ATCC_199 YFPSKVHLLVSALEREFEQIDAKSDRNAVSGG-TPYERLHAAITRLNRNM
MUL_4956|M.ulcerans_Agy99 YFSSKEHLLAAALQDWQMRLTRRILSASGSPDQDPLPGILDYLRRALRAF
MMAR_0141|M.marinum_M YFSSKEHLLAAALQDWQMRLTRRILSASGSPDQDPLPGILDYLRRALRAF
**.** ***.:** :: : : . : : : * :
Mb3605|M.bovis_AF2122/97 QRNPLLTEAMTRAYVFADASAASEVDQVEKLIDSMFARAMA-NGEPTEDQ
Rv3574|M.tuberculosis_H37Rv QRNPLLTEAMTRAYVFADASAASEVDQVEKLIDSMFARAMA-NGEPTEDQ
MAV_0584|M.avium_104 QRNPLLTEAMTRAYVFADASAASEVDQVEKLIDSMFARAMA-DGEPTEDQ
Mflv_1459|M.gilvum_PYR-GCK QRNPLLTEAMTRAFVFADASAAGEVDHVGKLMDSMFARAMS-DGEPTEDQ
Mvan_5312|M.vanbaalenii_PYR-1 QRNPLLTEAMTRAFVFADASAAGEVDHVGKLMDSMFARAMS-DGEPTEDQ
MSMEG_6042|M.smegmatis_MC2_155 QRNPLLTEAMTRAFVFADASAAGEVDHVGKLMDSMFARAMS-DGEPTEDQ
TH_1745|M.thermoresistible__bu QRNPLLTEAMTRAFVFADASAAGEVDHVGRLMDSMFARAMA-DGEPTEDQ
MAB_0579c|M.abscessus_ATCC_199 QRNPLLTEAMTRALVFADASAAGEVDHVGRLMDGIFARAMAGEDDPTDAQ
MUL_4956|M.ulcerans_Agy99 HRNPEMTALMLQMMTSTDRDAKATIDQMDRTNVEMFNRLLH--GVAPADI
MMAR_0141|M.marinum_M HRNPEMTALMLQMMTSTDRDAKATIDQMDRTNVEMFNRLLH--GVAPADI
:*** :* * : . :* .* . :*:: : :* * : . ..
Mb3605|M.bovis_AF2122/97 YHIARVISDVWLSNLLAWLTRRASATDVSKRLDLAVRLLIGDQDSA--
Rv3574|M.tuberculosis_H37Rv YHIARVISDVWLSNLLAWLTRRASATDVSKRLDLAVRLLIGDQDSA--
MAV_0584|M.avium_104 YHIARVISDVWLSNLLAWLTRRASATDVSKRLDLAVRLLLGEHG----
Mflv_1459|M.gilvum_PYR-GCK YHIARVISDVWLSNLLAWLTRRASATDVAKRLDLAVRLLIGDGDQPKI
Mvan_5312|M.vanbaalenii_PYR-1 YHIARVISDVWLSNLLAWLTRRASATDVAKRLDLAVRLLIGDGDQPKI
MSMEG_6042|M.smegmatis_MC2_155 YHIARVISDVWLSNLLAWLTRRASATDVSKRLDLAVRLLIGTEEQPKI
TH_1745|M.thermoresistible__bu YHIARVISDVWLSNLLAWLTRRASATDVSKRLELAVRLLVGDGKNPKV
MAB_0579c|M.abscessus_ATCC_199 FHIARVISDVWTSNMIAWLTRRASATDVSHRLDRTVRLLLGIT-----
MUL_4956|M.ulcerans_Agy99 SNVSFALNAALSGALTGLLTSRLTLEEALGHVEWVARALLDEGRPHP-
MMAR_0141|M.marinum_M SNVSFALNAALSGALTGLLTSRLTLEEALGHVEWVARALLDEGRPHP-
::: .:. . . : . ** * : :. ::: ..* *:.