For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARGLVIGEALIDIVDGPDPAEYVGGSPLNVAVGPARLGRDVDLLTHIGRDARGRRIAEYIESSGVQLVS GSQTADRTPTATARIGPDGSATYAFDLEWQIPDTPPVAPPLLVHTGSIAAAREPGCLAVAALLDAYRAAA TVSFDPNVRPSLRADPDLTRERIQRLVERSDIIKASAEDLHWIDPTQPPEQTARAWLACGPAIVALTFGD QSAVAFCAAGPASMPAQPVRVVDTVGAGDAFMAGLLDTLWEQGLLDADRRTELRKIGVSALTSALEVAAL TSALTVARAGADLPYRADLRQSR
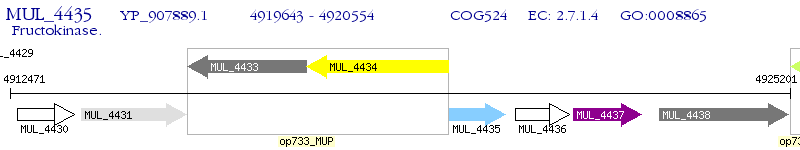
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4435 | pfkB | - | 100% (303) | fructokinase, PfkB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2462 | rbsK | 2e-09 | 27.39% (314) | ribokinase RBSK |
| M. gilvum PYR-GCK | Mflv_1827 | - | 1e-107 | 65.32% (297) | ribokinase-like domain-containing protein |
| M. tuberculosis H37Rv | Rv2436 | rbsK | 6e-09 | 28.15% (270) | ribokinase RBSK |
| M. leprae Br4923 | MLBr_02091 | - | 4e-07 | 52.17% (46) | hypothetical protein MLBr_02091 |
| M. abscessus ATCC 19977 | MAB_0359 | - | 4e-26 | 30.87% (311) | putative fructokinase |
| M. marinum M | MMAR_4574 | pfkB | 1e-167 | 98.02% (303) | fructokinase, PfkB |
| M. avium 104 | MAV_1051 | - | 1e-112 | 67.68% (297) | kinase, PfkB family protein |
| M. smegmatis MC2 155 | MSMEG_5577 | - | 1e-100 | 60.00% (305) | fructokinase |
| M. thermoresistible (build 8) | TH_0601 | - | 1e-104 | 63.42% (298) | fructokinase |
| M. vanbaalenii PYR-1 | Mvan_4916 | - | 1e-105 | 64.75% (295) | ribokinase-like domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4435|M.ulcerans_Agy99 ------------MARGLVIGEALIDIVDGP--DPA-----------EYVG
MMAR_4574|M.marinum_M ------------MARGLVIGEALIDIVDGP--DPA-----------EYVG
MAV_1051|M.avium_104 ------------MTRGLVIGESLIDIVED-----A-----------EYVG
Mflv_1827|M.gilvum_PYR-GCK -------------MRALVIGEALIDIVERGDEVT------------EHVG
Mvan_4916|M.vanbaalenii_PYR-1 -------------MRALVIGEALIDIVERDGGVT------------EHVG
MSMEG_5577|M.smegmatis_MC2_155 ------------MSRALVIGEALIDIVEREGQVVG-----------EHVG
TH_0601|M.thermoresistible__bu -----------MTARALVIGEALIDVVERDGRVLG-----------EHVG
MLBr_02091|M.leprae_Br4923 --------------------------------------------------
MAB_0359|M.abscessus_ATCC_1997 ---------MADGGGILVCGETLVDLVPVAGGLRR-----------PVPG
Mb2462|M.bovis_AF2122/97 MANASETNVGPMAPRVCVVGSVNMDLTFVVDALPRPGETVLAASLTRTPG
Rv2436|M.tuberculosis_H37Rv MANASETNVGPMAPRVCVVGSVNMDLTFVVDALPRPGETVLAASLTRTPG
MUL_4435|M.ulcerans_Agy99 GSPLNVAVGPARLGRDVDLLTHIGRDARGRRIAEYIESSGVQLVSGSQTA
MMAR_4574|M.marinum_M GSPLNVAVGLARLGRDVDLLTHIGRDARGRRIAEYIESSGVQLVSGSQTA
MAV_1051|M.avium_104 GSPLNVAVGLGRLGRDVDFLTYLADDDHGRRIAAYLKDAGVQLVSESRAA
Mflv_1827|M.gilvum_PYR-GCK GSPLNVAVGLARLGRDVDFLTHIGTDDRGRRIAEYVESSGVHLVPGSTAA
Mvan_4916|M.vanbaalenii_PYR-1 GSPLNVAVGLARLGRDVDFLTHIGTDARGQRITDFVESSGAQLVPGSRSA
MSMEG_5577|M.smegmatis_MC2_155 GSPLNVAVGLGRLGRDVDFLTHIADDPRGRRIVDYLKASGVQLVPGSTSA
TH_0601|M.thermoresistible__bu GSPLNVAVGLARLGRDVDFLTHIADDARGRRIADHLADSGVQLVPGSRTA
MLBr_02091|M.leprae_Br4923 --------------------------------------------------
MAB_0359|M.abscessus_ATCC_1997 GGPYNTAIAAAKLGAPTALLTHVSHDAFGSRCVQDLAAAGVDESLVMRHD
Mb2462|M.bovis_AF2122/97 GKGANQAVAAARAGAQVQFSGAFGDDPAAAQLRAHLRANAVGLDRTVTVP
Rv2436|M.tuberculosis_H37Rv GKGANQAVAAARAGAQVQFSGAFGDDPAAAQLRAHLRANAVGLDRTVTVP
MUL_4435|M.ulcerans_Agy99 DRTPTATARIGPDGSATYAFDLEWQ------IPDTPPVAPPLLVHTGSIA
MMAR_4574|M.marinum_M DRTPTATARIGPDGSATYAFDLEWQ------IPDTPPVAPPLLVHTGSIA
MAV_1051|M.avium_104 ERTATARSTIAADGSADYEFDLDWR------LSGIPPVAPPLFVHTGSIA
Mflv_1827|M.gilvum_PYR-GCK PRTATARAKLDAAGSASYSFNIEWA------LTGTPEVAPPLVVHTGSIA
Mvan_4916|M.vanbaalenii_PYR-1 ARTATARAVLDATGSATYSFDIDWQ------LSGTPEVAPPLVVHTGSIA
MSMEG_5577|M.smegmatis_MC2_155 KKTPTAQVRLDAAGSATYEFDIDWQ------LAGTAEVAPPIVAHSGSIA
TH_0601|M.thermoresistible__bu KRTPTAVARLDATGAADYTFDIDWQ------LSGTPEVAPPVVAHTGSIA
MLBr_02091|M.leprae_Br4923 --------------------------------------------------
MAB_0359|M.abscessus_ATCC_1997 VPTTLAVADVDEHGAARYLFYWQGTSNDVAIQSFPDPVRTPVAIWAGSIA
Mb2462|M.bovis_AF2122/97 GPSGTAIIVVDASAENTVLVAPGAN-------AHLTPVPSAVANCDVLLT
Rv2436|M.tuberculosis_H37Rv GPSGTAIIVVDASAENTVLVAPGAN-------AHLTPVPSAVANCDVLLT
MUL_4435|M.ulcerans_Agy99 AAREPGCLAVAALLDAYRAAATVSFDPNVRPSLRADPDLTRERIQRLVER
MMAR_4574|M.marinum_M AAREPGCLAVAALLDAYRAAATVSFDPNVRPSLSADPDLTRERIQRLVER
MAV_1051|M.avium_104 AVRDPGCLAVAALIDTYRVSATVTFDPNVRPSLIADRELAVSRIEHLVER
Mflv_1827|M.gilvum_PYR-GCK AVHEPGCLATKALIDAYHLSATFTFDPNIRAALITDADAARSRIDRLLEQ
Mvan_4916|M.vanbaalenii_PYR-1 AVLEPGCLATTALVDTYHLSATFTFDPNVRPALITDPDAARSRIDRLLER
MSMEG_5577|M.smegmatis_MC2_155 TVLEPGCRAAAALLDAYHLSATITFDPNVRPALISDGETARRRIERLVER
TH_0601|M.thermoresistible__bu AVLEPGCRAAAALIDTYHVSATTTFDPNVRPALITDDTGARARIDRFIER
MLBr_02091|M.leprae_Br4923 -----------------------------------------------MNR
MAB_0359|M.abscessus_ATCC_1997 SVLWRERDELRAWIARHYADIPLTFDVNVRPALISHRQTYAARIVPWLSV
Mb2462|M.bovis_AF2122/97 QLEIPVATALAAARAAQSADAVVMVN-------ASPAGQDRSSLQDLAAI
Rv2436|M.tuberculosis_H37Rv QLEIPVATALAAARAAQSADAVVMVN-------ASPAGQDRSSLQDLAAI
MUL_4435|M.ulcerans_Agy99 SDIIKASAEDLHWIDPTQPPEQTARAWLACGP--AIVALTFGDQSAVAFC
MMAR_4574|M.marinum_M SDIIKASAEDLHWIDPTQPPEQTARAWLACGP--AIVALTLGDQGAVAFC
MAV_1051|M.avium_104 SDIVKVSAEDLHWIDPDRSPEQLAQTWLGLGP--AVVAVTMADRGAVGYC
Mflv_1827|M.gilvum_PYR-GCK ADVVKASHEDLHWIDPTRTPEQVAAAWLALGP--AIVVVTFGADGALAMC
Mvan_4916|M.vanbaalenii_PYR-1 ADVVKASSEDLHWIDPTRNPEQVATAWLALGP--SIVVVTFGAEGSVAMC
MSMEG_5577|M.smegmatis_MC2_155 ADVVKISDEDLRWLDSRRSPEQIAKTWLSMGP--SIVAVTAGGEGSFAVC
TH_0601|M.thermoresistible__bu CDVVKASDEDLQWLEPGRSPEQVAQTWLRMGP--SLVAVTMGDRGAYAVC
MLBr_02091|M.leprae_Br4923 PDLVDIWTGDR-----------------------------CGEQGALALC
MAB_0359|M.abscessus_ATCC_1997 ARVARASTDDLEFLYPGVSVRDAVNAWLDAYPRIEIAVITCGAAGSLAFR
Mb2462|M.bovis_AF2122/97 ADVVIAN-------------EHEANDWPSPPT---HFVITLGVRGARYVG
Rv2436|M.tuberculosis_H37Rv ADVVIAN-------------EHEANDWPSPPT---HFVITLGVRGARYVG
: . .:
MUL_4435|M.ulcerans_Agy99 AAG--PASMPAQPVRVVDTVGAGDAFMAGLLDTLWEQGLLDADRRTELRK
MMAR_4574|M.marinum_M AAG--PASVPAQPVRVVDTVGAGDAFMAGLLDTLWEQGLLGADRRTELRK
MAV_1051|M.avium_104 AAG--TARVPTRAVRVVDTVGAGDSFMAGLLDALWEAGLLGGDRRGALRD
Mflv_1827|M.gilvum_PYR-GCK AAG--SVRVPAGNVHVVDTVGAGDAFMTGLIDALWSADLLGADRRHHLAR
Mvan_4916|M.vanbaalenii_PYR-1 ADG--TVRVPAGNVDVVDTVGAGDAFMTGLIDALWSMDLLGAGRRARLAG
MSMEG_5577|M.smegmatis_MC2_155 GAG--MVRVPARTVEVIDTVGAGDAFMTGLLDALWSLDLLGADRRRNLAH
TH_0601|M.thermoresistible__bu AAG--EVRVPARPVEVVDTVGAGDAFMAGLIDALWSLQLLGAERRRQLAG
MLBr_02091|M.leprae_Br4923 LAG--KARVDIRNFRVVDTVCTSDEFMVRLLDGLWGYGL-----------
MAB_0359|M.abscessus_ATCC_1997 RGEPSPLRIAAHKVSVVDTVGAGDTYTGAFLDGFYRHRLD----------
Mb2462|M.bovis_AF2122/97 ADG--VFEVPAPTVTPVDTAGAGDVFAG-VLAANWPRNPGSPAER-----
Rv2436|M.tuberculosis_H37Rv ADG--VFEVPAPTVTPVDTAGAGDVFAG-VLAANWPRNPGSPAER-----
: . :**. :.* : .: :
MUL_4435|M.ulcerans_Agy99 IGVSALTSALEVAALTSALTVARAGADLPY---RADLRQSR-------
MMAR_4574|M.marinum_M IGVSALTSALEVAALTSALTVARAGADLPY---RADLRQSR-------
MAV_1051|M.avium_104 IGIDTLTSALDAASLSSALTIARAGADLPD---RAAVQAALRR-----
Mflv_1827|M.gilvum_PYR-GCK IDVDALTTAVRAATLTSALTVAKSGADLPDRVERDTAPASDILS----
Mvan_4916|M.vanbaalenii_PYR-1 IGTDALTTAVRAATLTAALTVTRPGADLPDRVTRDAATASDILG----
MSMEG_5577|M.smegmatis_MC2_155 ISTDALEHVLSTAALTSALTVAKAGADLPD---RDALRAAQQ------
TH_0601|M.thermoresistible__bu IAVEALQRVVETAALSSALTVARAGADLPDRATRDAAAAATERETGTY
MLBr_02091|M.leprae_Br4923 ------------------------------------------------
MAB_0359|M.abscessus_ATCC_1997 -----LPEALRRAAVAAAITCTRPGAQPPDATELANELRRVTS-----
Mb2462|M.bovis_AF2122/97 --LRALRRACAAGVLATLVSGAGDCAPAAAAIDAALRANRHNGS----
Rv2436|M.tuberculosis_H37Rv --LRALRRACAAGALATLVSGVGDCAPAAAAIDAALRANRHNGS----