For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGETYESVTVEIKDQVAQATLIGPGKGNAMGPAFWSELPEVFAALDADRDVRAIVITGSGKNFSYGLDVP AMGGTFAPLMAEGALARPRTDFHAEVLRMQKATNAVADCRTPTIASVHGWCIGGGVDLISAVDMRYASAD AKFSIREVKLAIVADMGSLARLPMILNDGHLRELALTGRDIDAARAEKIGLVNQVFEDADATLAAAHATA TEIAANPPLAVYGVKDVLDQQRASAVAENLRYVAAWNAAFLPSKDLTEGISATFAKRPPQFTGEQAAAST YPRGVASMTEDGRIRVPADLDAVTALGAEDHSEIDSAAIERIWQATRYWYQAGMHPAIQLCIRHNGRVVL NRAIGHGWGNAPSDAPDAEKIPVSTDTPFCTYSSAKAITATVVHMLAERGHFSLDDRVCEYIPSYTSHGK DHTTIRHVLTHSAGVPFPTGPRPDVTRADDHDYAARKLGELRPLYPPGLVHIYHALTWGPLMREIIWATT RKEIRDILATEILDPLGFRWTNFGVAEEDLPLVAPSHATGRPLPPVIAAAFRKAIGGTVHEVIPYTNKPL FLRTIIPSSNTVSTANELSRFMEILRRGGELDGVRIMAPETVRSAVTECRRLRPDSATGLAPLRWGTGFM LGSNRFGPFGRNAPAAFGHLGLVNIATWADPDRGLSVGLINSGKPGRDPDAKRYVALKNAITAEIPPKTV E
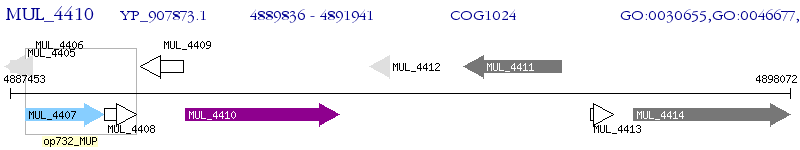
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4410 | - | - | 100% (701) | fusion of enoyl-CoA hydratase, EchA21 and lipase, LipE |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 5e-32 | 35.19% (270) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_1114 | echA1 | 3e-27 | 32.48% (274) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3804 | lipE | 0.0 | 84.98% (406) | putative lipase LIPE |
| M. gilvum PYR-GCK | Mflv_1067 | - | 1e-142 | 65.29% (363) | beta-lactamase |
| M. tuberculosis H37Rv | Rv3775 | lipE | 0.0 | 84.98% (406) | lipase LipE |
| M. leprae Br4923 | MLBr_00119 | lipE | 0.0 | 81.91% (409) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0064c | - | 0.0 | 72.59% (405) | lipase LipE |
| M. marinum M | MMAR_5330 | lipE | 0.0 | 98.31% (414) | lipase LipE |
| M. avium 104 | MAV_0246 | - | 0.0 | 77.43% (412) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_6575 | - | 1e-171 | 67.73% (406) | beta-lactamase |
| M. thermoresistible (build 8) | TH_2342 | lipE | 0.0 | 72.51% (411) | PROBABLE LIPASE LIPE |
| M. vanbaalenii PYR-1 | Mvan_5757 | - | 1e-148 | 65.58% (369) | beta-lactamase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4410|M.ulcerans_Agy99 MGETYESVTVEIKDQVAQATLIGPGKGNAMGPAFWSELPEVFAALDADRD
MMAR_5330|M.marinum_M --------------------------------------------------
Mb3804|M.bovis_AF2122/97 --------------------------------------------------
Rv3775|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00119|M.leprae_Br4923 --------------------------------------------------
MAV_0246|M.avium_104 --------------------------------------------------
Mflv_1067|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2342|M.thermoresistible__bu --------------------------------------------------
MSMEG_6575|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0064c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 VRAIVITGSGKNFSYGLDVPAMGGTFAPLMAEGALARPRTDFHAEVLRMQ
MMAR_5330|M.marinum_M --------------------------------------------------
Mb3804|M.bovis_AF2122/97 --------------------------------------------------
Rv3775|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00119|M.leprae_Br4923 --------------------------------------------------
MAV_0246|M.avium_104 --------------------------------------------------
Mflv_1067|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2342|M.thermoresistible__bu --------------------------------------------------
MSMEG_6575|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0064c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 KATNAVADCRTPTIASVHGWCIGGGVDLISAVDMRYASADAKFSIREVKL
MMAR_5330|M.marinum_M --------------------------------------------------
Mb3804|M.bovis_AF2122/97 --------------------------------------------------
Rv3775|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00119|M.leprae_Br4923 --------------------------------------------------
MAV_0246|M.avium_104 --------------------------------------------------
Mflv_1067|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2342|M.thermoresistible__bu --------------------------------------------------
MSMEG_6575|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0064c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 AIVADMGSLARLPMILNDGHLRELALTGRDIDAARAEKIGLVNQVFEDAD
MMAR_5330|M.marinum_M --------------------------------------------------
Mb3804|M.bovis_AF2122/97 --------------------------------------------------
Rv3775|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00119|M.leprae_Br4923 --------------------------------------------------
MAV_0246|M.avium_104 --------------------------------------------------
Mflv_1067|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2342|M.thermoresistible__bu --------------------------------------------------
MSMEG_6575|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0064c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 ATLAAAHATATEIAANPPLAVYGVKDVLDQQRASAVAENLRYVAAWNAAF
MMAR_5330|M.marinum_M --------------------------------------------------
Mb3804|M.bovis_AF2122/97 --------------------------------------------------
Rv3775|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00119|M.leprae_Br4923 --------------------------------------------------
MAV_0246|M.avium_104 --------------------------------------------------
Mflv_1067|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2342|M.thermoresistible__bu --------------------------------------------------
MSMEG_6575|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0064c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 LPSKDLTEGISATFAKRPPQFTGEQAAASTYPRGVASMTEDGRIRVPADL
MMAR_5330|M.marinum_M -------------------------------------MTEDGRIRVPADL
Mb3804|M.bovis_AF2122/97 ------------------------------------MRAGDGKIRVPADL
Rv3775|M.tuberculosis_H37Rv ------------------------------------MRAGDGKIRVPADL
MLBr_00119|M.leprae_Br4923 ----------------------------------MTS---DGKIRVPTDL
MAV_0246|M.avium_104 ----------------------------------MTTPTLDGKIRLPADL
Mflv_1067|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2342|M.thermoresistible__bu ---------------------------MTRDNTTGAGATPDGRIRVPADL
MSMEG_6575|M.smegmatis_MC2_155 ---------------------------------------MTGKISVPADL
MAB_0064c|M.abscessus_ATCC_199 ----------------------------------MTASDQAGRIRVPADL
MUL_4410|M.ulcerans_Agy99 DAVTALGAEDHSEIDSAAIERIWQATRYWYQAGMHPAIQLCIRHNGRVVL
MMAR_5330|M.marinum_M DAVTALGAEDHSEIDSAAIERIWQAARYWYQAGMHPAIQLCIRHNGRVVL
Mb3804|M.bovis_AF2122/97 DAVTATGEEDHSEIDGAAVDRIWRAARHWYRAGMHPAIQLCIRHHGRVVL
Rv3775|M.tuberculosis_H37Rv DAVTATGEEDHSEIDGAAVDRIWRAARHWYRAGMHPAIQLCIRHHGRVVL
MLBr_00119|M.leprae_Br4923 DAVTAIGDEDHSDIDSAAVERIWQATRHWYQAGMHPAIQLCIRQHGRVVL
MAV_0246|M.avium_104 DAVTTIGAEDHSEIDAAAVDRIWAAGRHWYQGGFHPAIQLCIRRHGRVVL
Mflv_1067|M.gilvum_PYR-GCK ---------------------------------MQPAIQVCLRRDGRVIL
Mvan_5757|M.vanbaalenii_PYR-1 --------------------------MHWYRAGMQPAIQVCLRRDGKVIL
TH_2342|M.thermoresistible__bu DAVTAVGDEDHTGIDPSAVERIWQAVRHWYRAGMHPAIQVCVRHDGRVVL
MSMEG_6575|M.smegmatis_MC2_155 DAVTTIGDEDHSGIDARAIEQIWDSVRYWYQSGLHPAIQVCLRHKGRVVL
MAB_0064c|M.abscessus_ATCC_199 DSVTDIAEEDHSDIGPAAVERIWQAARYWYQAGMHPAIQVCLRHRGKVIL
::****:*:*: *:*:*
MUL_4410|M.ulcerans_Agy99 NRAIGHGWGNAPSDAPDAEKIPVSTDTPFCTYSSAKAITATVVHMLAERG
MMAR_5330|M.marinum_M NRAIGHGWGNAPSDAPDAEKIPVSTDTPFCTYSSAKAITATVAHMLAERG
Mb3804|M.bovis_AF2122/97 NRAIGHGWGNAPTDEADAEKIPVTTDTPFCVYSAAKAITATVVHMLVERG
Rv3775|M.tuberculosis_H37Rv NRAIGHGWGNAPTDEADAEKIPVTTDTPFCVYSAAKAITATVVHMLVERG
MLBr_00119|M.leprae_Br4923 NRAIGHGWGNAPTDPHDAEKIPVTTDTPFCAYSAAKAITATVVHMLVERG
MAV_0246|M.avium_104 NRAIGHGWGNAPTDPPDAEKIPVTPDTPFCVYSAAKGIAATVVHMLVERG
Mflv_1067|M.gilvum_PYR-GCK DRAIGYAA----------DDVPVRTDTPFCVYSAAKAITTTVTHMLVERG
Mvan_5757|M.vanbaalenii_PYR-1 DRAIGYAW----------DDVPVRTDTPFCVYSAAKAITTTVTHMLVERG
TH_2342|M.thermoresistible__bu NRAIGHGWGNAPTDPPDAEKVPVRVDTPFCVYSAAKAIATTVVHMLVERG
MSMEG_6575|M.smegmatis_MC2_155 NRAIGHGWGNGPDDSPDTEKILATPETPFCVYSAAKAISSTVVHMLVERG
MAB_0064c|M.abscessus_ATCC_199 NRAIGHGWGNAPTDPVDAEKIPVTTDTPFCVYSAAKAISTTVVHLLVERG
:****:. :.: . :****.**:**.*::**.*:*.***
MUL_4410|M.ulcerans_Agy99 HFSLDDRVCEYIPSYTSHGKDHTTIRHVLTHSAGVPFPTGPRPDVTRADD
MMAR_5330|M.marinum_M HFSLDDRVCEYIPSYTSHGKDRTTIRHVLTHSAGVPFPTGPRPDVTRADD
Mb3804|M.bovis_AF2122/97 HFALDDRVCEYLPSYTSHGKHRTTIRHVLTHSAGVPFPTGPRPDVRRADD
Rv3775|M.tuberculosis_H37Rv HFALDDRVCEYLPSYTSHGKHRTTIRHVLTHSAGVPFPTGPRPDVRRADD
MLBr_00119|M.leprae_Br4923 HFSLDDRVCEYLPTYTSYGKHRTTIRHVLTHSAGVPFPTGPRPDVTRADD
MAV_0246|M.avium_104 VFSLDDRVCDYIPTFTSHGKHRITIRHVMTHSAGLPFPTGPRPDVTRADD
Mflv_1067|M.gilvum_PYR-GCK EFSLDDRVCEYLPEYTSHGKDRTTIRHVLTHSAGIPFATGPKPDLKRMDD
Mvan_5757|M.vanbaalenii_PYR-1 AFSLEDRVCDYLPEYTSHGKDRTTIRHVLTHSAGIPFATGPKPDLKRMDD
TH_2342|M.thermoresistible__bu HLSLDDRVCDHLPGYTSHGKDRTTIRHVLTHSAGVPFATGPRPDLRRMDD
MSMEG_6575|M.smegmatis_MC2_155 YFSLDDRVCDYLPEYTSHGKDRTTIRHVMTHSAGVPIHTGPRPDVTKVND
MAB_0064c|M.abscessus_ATCC_199 ALSLDDRVCEYLPTFTSRGKDRITIRHVMTHSAGIPVPTGPRPDITRMDD
::*:****:::* :** **.: *****:*****:*. ***:**: : :*
MUL_4410|M.ulcerans_Agy99 HDYAARKLGELRPLYPPGLVHIYHALTWGPLMREIIWATTRKEIRDILAT
MMAR_5330|M.marinum_M HEYAARKLGELRPLYPPGLVHIYHALTWGPLMREIIWATTGKEIRDILAT
Mb3804|M.bovis_AF2122/97 HEYAVERLGELRPLYRPGLVHIYHALTWGPLMREIVYAATGKEIREILAT
Rv3775|M.tuberculosis_H37Rv HEYAVERLGELRPLYRPGLVHIYHALTWGPLMREIVYAATGKEIREILAT
MLBr_00119|M.leprae_Br4923 HAYAQQKLSELRPFYRPGLVHIYHALTWGPLMREIIWTATGKEIRDILAA
MAV_0246|M.avium_104 HEYAQQKLGELRPLYRPGLFHMYHALTWGPLVREIVYAATGKEIREILAT
Mflv_1067|M.gilvum_PYR-GCK SDYTRAMLARMKPVYRPGLIHVYHGVTWGPLMREIISAATGRSIRDILAD
Mvan_5757|M.vanbaalenii_PYR-1 SAYTREMLGRMKPVYPPGLVHMYHGVTWGPLMREIISAATGRSIRDILAE
TH_2342|M.thermoresistible__bu SEYARTMLGNLKPLYRPGLVHIYHGVTWGPLIREIVSAATGRNIRDILAT
MSMEG_6575|M.smegmatis_MC2_155 SEYTRQQLAALKPVYRPGLIHIYHGLTWGPLIREIVSAATGRNIRDILAE
MAB_0064c|M.abscessus_ATCC_199 SEYTREQLGQLRPLYRPGLVHMYHALTWGPLVREIVLGATGKGIRDILAT
*: *. ::*.* ***.*:**.:*****:***: :* : **:***
MUL_4410|M.ulcerans_Agy99 EILDPLGFRWTNFGVAEEDLPLVAPSHATGRPLPPVIAAAFRKAIGGTVH
MMAR_5330|M.marinum_M EILDPLGFRWTNFGVAEEDLPLVAPSHATGRPLPPVIAAAFRKAIGGTVH
Mb3804|M.bovis_AF2122/97 EILDPLGFRWTNFGVAERDVPLVAPSHATGRQLPPVIAAVFRKAIGGTVH
Rv3775|M.tuberculosis_H37Rv EILDPLGFRWTNFGVAERDVPLVAPSHATGRQLPPVIAAVFRKAIGGTVH
MLBr_00119|M.leprae_Br4923 EILDPLGFRWTNFGVAEEDVPLVALSHATGRPLPPIIAAIFRKAIGGTVH
MAV_0246|M.avium_104 EILDPLGFRWTNFGVAKQDVPLVAPSHATGRTLPPVVAQIFRKAIGGTVH
Mflv_1067|M.gilvum_PYR-GCK EILTPLGFRWTNYGVAPEDVPLVARSHVTGKPLPAPVAKAFKAAVGGTPQ
Mvan_5757|M.vanbaalenii_PYR-1 EILTPLGFRWTNYGVAAQDVPLVAPSHVTGKPLPAPIAKAFKTAVGGTPQ
TH_2342|M.thermoresistible__bu EILEPLGFRWTNFGVAPDDVPLVAPSHPTGKPLPAPIAAAFRTAVGGTLH
MSMEG_6575|M.smegmatis_MC2_155 EILEPLKFRWTNYGVRPEEVDLVAPSHVTGKPLPAPIAAAFRAAVGGTPD
MAB_0064c|M.abscessus_ATCC_199 EILDPLGFRWTNYGVAPEDVPLVAPSHPTGKPLPAPMRAAFRAVVGGTMH
*** ** *****:** :: *** ** **: **. : *: .:*** .
MUL_4410|M.ulcerans_Agy99 EVIPYTNKPLFLRTIIPSSNTVSTANELSRFMEILRRGGELDGVRIMAPE
MMAR_5330|M.marinum_M EVIPYTNKPLFLRTIIPSSNTVSTANELSRFMEILRRGGELDGVRIMAPE
Mb3804|M.bovis_AF2122/97 EIIPYTNTPFFLSTILPSSNTVSTANELSRFMEILRRGGELDGVRVLSPE
Rv3775|M.tuberculosis_H37Rv EIIPYTNTPFFLSTILPSSNTVSTANELSRFMEILRRGGELDGVRVLSPE
MLBr_00119|M.leprae_Br4923 EVIPYTNTPPFLTTIVPSSNTVSTANELSRFAEIWLRGGELDGIRVLRSE
MAV_0246|M.avium_104 EMIPYTNSPMFLTTIIPSSNTVSTAHEMSRFAEIWRRGGELDGVRVISPE
Mflv_1067|M.gilvum_PYR-GCK QIIPFSNTRQFLTGVIPSSNTVSNAHELSRFAEILCRGGELDGVRVLSAE
Mvan_5757|M.vanbaalenii_PYR-1 QIIPFSNTREFLTGVIPSSNTVSNANELSRFAEILCRGGELDGIRVMSPE
TH_2342|M.thermoresistible__bu QIIPFSNTPQFLTGVVPSSNTVSNAFELSRFAELLRRGGELDGVRVLAPE
MSMEG_6575|M.smegmatis_MC2_155 RIIPLSNTPLFLTSIVPSSSTVSNADELSRFAEILRRGGELDGVRILRPE
MAB_0064c|M.abscessus_ATCC_199 EIIPMSNQPFFLTGVVPSSNTVSTANELSRFAEILCRGGELDGVRVMSPE
.:** :* ** ::***.***.* *:*** *: *******:*:: .*
MUL_4410|M.ulcerans_Agy99 TVRSAVTECRRLRPDSATGLAPLRWGTGFMLGSNRFGPFGRNAPAAFGHL
MMAR_5330|M.marinum_M TVRSAATECRRLRPDFATGLAPLRWGTGFMLGSNRFGPFGRNAPAAFGHL
Mb3804|M.bovis_AF2122/97 TLRGAVTECRRLRPDFATGLMPLRWGTGFMLGSAKYGPFGRNAPAAFGHL
Rv3775|M.tuberculosis_H37Rv TLRGAVTECRRLRPDFATGLMPLRWGTGFMLGSAKYGPFGRNAPAAFGHL
MLBr_00119|M.leprae_Br4923 TLLGAVAECRRLRPDFVVGLMPARWGTGFILGTNKFGPFGRNAPAAFGHL
MAV_0246|M.avium_104 TMYHAVAECRRLRPDFAVGLQPARWGTGFILGTNRWGPFGRNAPHAFGSL
Mflv_1067|M.gilvum_PYR-GCK TLRAATKEARRLRPDLATGLQPMRWGTGYMLGSKRFGPFGRGADGAFGHT
Mvan_5757|M.vanbaalenii_PYR-1 TLRAATKEARRLRPDLATGLQPMRWGTGYMLGSKRFGPFGRDAAGAFGHT
TH_2342|M.thermoresistible__bu TLRAATRQCRRLRPDLATGGQPLRWGLGYMLGSKRFGPFGRNAPAAFGHT
MSMEG_6575|M.smegmatis_MC2_155 TLRAATAPCRRIRPDVATGLKPMRWGTGYMLGGKRFGPFGRNTESAFGHT
MAB_0064c|M.abscessus_ATCC_199 TIRAAAAPARRLRPDMATGGMPMRWGTGYMLGSKRFGPFGRNSPAAFGHT
*: *. .**:*** ..* * *** *::** ::*****.: ***
MUL_4410|M.ulcerans_Agy99 GLVNIATWADPDRGLSVGLINSGKPGRDPDAKRYVALKNAITAEIPPKTV
MMAR_5330|M.marinum_M GLVNIATWADPDRGLSVGLINSGKPGRDPDAKRYVALKNAITAEIPPKTV
Mb3804|M.bovis_AF2122/97 GLVNIAVWADPERALSGGLISSGKPGRDPEAGRYGALLNAITAEIPRASS
Rv3775|M.tuberculosis_H37Rv GLVNIAVWADPERALSGGLISSGKPGRDPEAGRYGALLNAITAEIPRASS
MLBr_00119|M.leprae_Br4923 GLVNIAIWADPQRSMAAGLISSGKPGQDPNAGRYGALMNTITAEIPVH--
MAV_0246|M.avium_104 GLTNIAIWADPARGLAAGVISSGKPGRDPEARRYTALMDTITAAIPRD--
Mflv_1067|M.gilvum_PYR-GCK GLTDIAVWADPQRALSVAVVSSGKPGPHREAKRYPRLLDRINAEIPRL--
Mvan_5757|M.vanbaalenii_PYR-1 GLTDIVVWADPQRALSVAVVSSGKPGSHREARRYPALLDRINAEIPRVS-
TH_2342|M.thermoresistible__bu GLVNIAMWADPERRLAAAVVSSGKPGNHREANRYPAVLDRIAAEFPRA--
MSMEG_6575|M.smegmatis_MC2_155 GLVDIAVWADPARSLSAAVVSSGKPGAHFETNRYPAVLNRIAEQIPPG--
MAB_0064c|M.abscessus_ATCC_199 GLVDIAVWADPARALSVGVVSSGKPGNHKEAKRYPALLDLIAAEVPLVG-
**.:*. **** * :: .::.***** . :: ** : : * .*
MUL_4410|M.ulcerans_Agy99 E
MMAR_5330|M.marinum_M E
Mb3804|M.bovis_AF2122/97 G
Rv3775|M.tuberculosis_H37Rv G
MLBr_00119|M.leprae_Br4923 -
MAV_0246|M.avium_104 -
Mflv_1067|M.gilvum_PYR-GCK -
Mvan_5757|M.vanbaalenii_PYR-1 -
TH_2342|M.thermoresistible__bu -
MSMEG_6575|M.smegmatis_MC2_155 -
MAB_0064c|M.abscessus_ATCC_199 -