For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LATSAEKSDMTHIAKPLTKSALAGGFVVASMSLSTGVAQADLMNWEAVAQCESGGNWSVNTGNGAYGGLQ FKQATWEEYGGVGNPASASKQQQIAIANRVLAGQGPAAWPKCSSAGGLPPVPVVLPKPVRQLEQTVTQVI SIFTPR
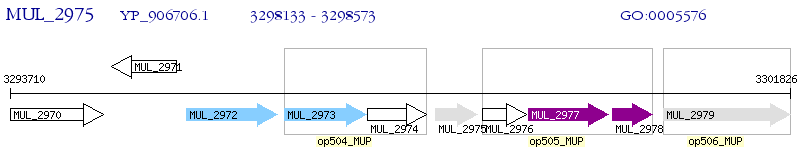
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2975 | - | - | 100% (146) | resuscitation-promoting factor-like protein |
| M. ulcerans Agy99 | MUL_3723 | rpfE | 2e-22 | 55.41% (74) | resuscitation-promoting factor RpfE |
| M. ulcerans Agy99 | MUL_0283 | rpfA | 3e-22 | 43.20% (125) | resuscitation-promoting factor RpfA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1916c | rpfC | 1e-45 | 62.86% (140) | resuscitation-promoting factor RpfC |
| M. gilvum PYR-GCK | Mflv_2620 | - | 5e-32 | 53.23% (124) | transglycosylase domain-containing protein |
| M. tuberculosis H37Rv | Rv1884c | rpfC | 1e-45 | 62.86% (140) | resuscitation-promoting factor RpfC |
| M. leprae Br4923 | MLBr_02030 | - | 1e-36 | 51.06% (141) | hypothetical protein MLBr_02030 |
| M. abscessus ATCC 19977 | MAB_4080c | - | 4e-33 | 66.67% (87) | hypothetical protein MAB_4080c |
| M. marinum M | MMAR_2772 | - | 3e-76 | 98.54% (137) | resuscitation-promoting factor-like protein |
| M. avium 104 | MAV_2818 | - | 1e-31 | 57.28% (103) | resuscitation-promoting factor RpfA |
| M. smegmatis MC2 155 | MSMEG_4640 | - | 2e-35 | 62.75% (102) | secreted protein |
| M. thermoresistible (build 8) | TH_0358 | - | 2e-32 | 53.33% (120) | PROBABLE RESUSCITATION-PROMOTING FACTOR RPFE |
| M. vanbaalenii PYR-1 | Mvan_3961 | - | 5e-32 | 49.21% (126) | transglycosylase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2975|M.ulcerans_Agy99 ---------------------------MATSAEKSDMTHIAKPLTKSALA
MMAR_2772|M.marinum_M ------------------------------------MTHIAKPLTKSALA
Mb1916c|M.bovis_AF2122/97 MHPLPADHGRSRCNRHPISPLSLIGNASATSGDMSSMTRIAKPLIKSAMA
Rv1884c|M.tuberculosis_H37Rv MHPLPADHGRSRCNRHPISPLSLIGNASATSGDMSSMTRIAKPLIKSAMA
MAV_2818|M.avium_104 --------------------------------------------------
MLBr_02030|M.leprae_Br4923 ---------------------------MDMPGEMLDVRKLCKLFVKSAVV
Mflv_2620|M.gilvum_PYR-GCK ----------------------------MN-----VRKLAGKGLMAAAIS
Mvan_3961|M.vanbaalenii_PYR-1 ----------------------------MN-----VRKVVSKGLMAAAIS
MSMEG_4640|M.smegmatis_MC2_155 ---------------------------------------MKRAFGLTALA
TH_0358|M.thermoresistible__bu ----------------------------MNNIRKALTSSLKRALLLIAGT
MAB_4080c|M.abscessus_ATCC_199 ----------------------------MDR-----ARWVRDRRAVCLIF
MUL_2975|M.ulcerans_Agy99 GGFVVASMSLST--GVAQADL--MNWEAVAQCESGGNWSVNTGNGAYGGL
MMAR_2772|M.marinum_M GGFVVASMSLST--GVAQADL--MNWEAVAQCESGGNWSANTGNGAYGGL
Mb1916c|M.bovis_AF2122/97 AGLVTASMSLST--AVAHAGP-SPNWDAVAQCESGGNWAANTGNGKYGGL
Rv1884c|M.tuberculosis_H37Rv AGLVTASMSLST--AVAHAGP-SPNWDAVAQCESGGNWAANTGNGKYGGL
MAV_2818|M.avium_104 ---------------MVSAEP-TPNWDAIAQCESGGNWHANTGNGEYGGL
MLBr_02030|M.leprae_Br4923 SGIVTASMALSTSTGMANAVPREPNWDAVAQCESGRNWRANTGNGFYGGL
Mflv_2620|M.gilvum_PYR-GCK GALAVVPMALDGGTATAHADS--VNWDAIAQCESGGNWAINTGNGHFGGL
Mvan_3961|M.vanbaalenii_PYR-1 GALAVVPMAMDGTTATAHADS--VNWDAIAQCESGGNWAINTGNGHFGGL
MSMEG_4640|M.smegmatis_MC2_155 GALAMGPMVLS--PATAAADS--VNWDAIAACESGGNWSTNTGNGHYGGL
TH_0358|M.thermoresistible__bu AALALVPMVLS--TATAQADT--VNWDAIAQCESGGNWASDTGNGFYGGL
MAB_4080c|M.abscessus_ATCC_199 TVLFMMSLSMT--ASVASADS--MNWDAVAECESGGNWSANTGNGFYGGL
. * **:*:* **** ** :**** :***
MUL_2975|M.ulcerans_Agy99 QFKQATWEEYGGVGNPASASKQQQIAIANRVLAGQGPAAWPKCSSAGGLP
MMAR_2772|M.marinum_M QFKQATWEEYGGVGNPASASKQQQIAIANRVLAGQGPAAWPKCSSAGGLP
Mb1916c|M.bovis_AF2122/97 QFKPATWAAFGGVGNPAAASREQQIAVANRVLAEQGLDAWPTCGAASGLP
Rv1884c|M.tuberculosis_H37Rv QFKPATWAAFGGVGNPAAASREQQIAVANRVLAEQGLDAWPTCGAASGLP
MAV_2818|M.avium_104 QFKPATWARYGGVGNPAAASREQQIAVANRVFAEEGVEPWPKCGAQSGLP
MLBr_02030|M.leprae_Br4923 QFKPTIWARYGGVGNPAGASREQQITVANRVLADQGLDAWPKCGAASDLP
Mflv_2620|M.gilvum_PYR-GCK QFKQATWSANGGVGNPARAPRHEQIRVAENVLRTQGIKAWPKCGAKGMAA
Mvan_3961|M.vanbaalenii_PYR-1 QFKQATWSANGGVGNPAAAPRHEQIRVAENVLRTQGLKAWPKCGAKGMAA
MSMEG_4640|M.smegmatis_MC2_155 QFKQSTWAAHGGVGSPATASREQQIAVAENVLRTQGLAAWPKCGARGGVP
TH_0358|M.thermoresistible__bu QFKPSTWAAHGGTGSPARASREEQIRVAERVLASQGLKAWPKCGAHGTRP
MAB_4080c|M.abscessus_ATCC_199 QFKPSTWAAHGGVGNPAEASREEQIAVAEDVLRTQGPGAWPKCGGGGIGG
*** : * **.*.** *.:.:** :*: *: :* .**.*.. .
MUL_2975|M.ulcerans_Agy99 PV----------------------PVVLPKPVRQLEQTVTQV------IS
MMAR_2772|M.marinum_M PV----------------------PVVLPKPVRQLEQTVTQI------IS
Mb1916c|M.bovis_AF2122/97 ------------------------IALWSKPAQGIKQIINEIIWAG--IQ
Rv1884c|M.tuberculosis_H37Rv ------------------------IALWSKPAQGIKQIINEIIWAG--IQ
MAV_2818|M.avium_104 ------------------------IGWYSHPAQGIKQIINGL------IQ
MLBr_02030|M.leprae_Br4923 ------------------------ITLWSHPAQGVKQIINDIIQMGDTTL
Mflv_2620|M.gilvum_PYR-GCK QVWGNPAPSAPVPAVTPAATPGRANGCATMPTSVFGGIVDLRKMCTALFV
Mvan_3961|M.vanbaalenii_PYR-1 QVWGNTAPTAPVPATVPAATPGRANGCATMPTSVFGGVLDLRKMCTALFT
MSMEG_4640|M.smegmatis_MC2_155 AVWGG-GAGAPAAG----------SGCSAVRPGAVLGILDLRRLCSTLLD
TH_0358|M.thermoresistible__bu AVFR--TPGAPAAGGVPASTPTATTGCGAIRPGAVLGLVDLRQVCAAFLT
MAB_4080c|M.abscessus_ATCC_199 --------G---------------AGCQFVPGGSIFGIVNFRQMCNGVMG
. :
MUL_2975|M.ulcerans_Agy99 IFTPR---
MMAR_2772|M.marinum_M IFTPR---
Mb1916c|M.bovis_AF2122/97 ASIPR---
Rv1884c|M.tuberculosis_H37Rv ASIPR---
MAV_2818|M.avium_104 AAVPR---
MLBr_02030|M.leprae_Br4923 AAIALNGL
Mflv_2620|M.gilvum_PYR-GCK PRSAR---
Mvan_3961|M.vanbaalenii_PYR-1 PRSAR---
MSMEG_4640|M.smegmatis_MC2_155 PLGAFGR-
TH_0358|M.thermoresistible__bu PLGVG---
MAB_4080c|M.abscessus_ATCC_199 LIPPAG--