For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VFKLLFYSPRIAPNTGNAIRTAAATGCELHLVEPLGFDLSEPKLRRAGLDYHDLASVTVHGSLTQAWDAL LPARVFAFTAGASVTFSDVDFRPGDVLMFGPEPTGLHAATLADAHITQQVRIPMLAGRRSLNLANAAAVA VYEAWRQHGYPGAL
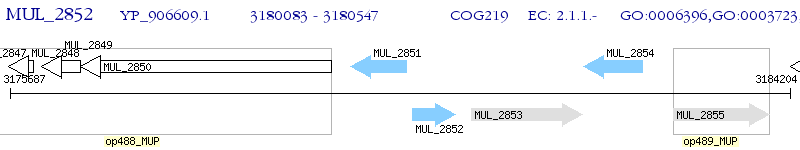
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2852 | spoU | - | 100% (154) | tRNA/rRNA methyltransferase SpoU |
| M. ulcerans Agy99 | MUL_1630 | tsnR | 3e-08 | 32.85% (137) | 23S rRNA methyltransferase TsnR |
| M. ulcerans Agy99 | MUL_0269 | - | 5e-05 | 34.27% (143) | rRNA methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3401 | spoU | 3e-71 | 83.12% (154) | tRNA/rRNA methylase SpoU |
| M. gilvum PYR-GCK | Mflv_4891 | - | 9e-69 | 79.87% (154) | tRNA/rRNA methyltransferase (SpoU) |
| M. tuberculosis H37Rv | Rv3366 | spoU | 3e-71 | 83.12% (154) | tRNA/rRNA methylase SpoU |
| M. leprae Br4923 | MLBr_00419 | - | 8e-70 | 80.92% (152) | putative tRNA/rRNA methyltransferase |
| M. abscessus ATCC 19977 | MAB_3702 | - | 1e-61 | 70.78% (154) | tRNA/rRNA methyltransferase SpoU |
| M. marinum M | MMAR_1163 | spoU | 2e-85 | 99.35% (154) | tRNA/rRNA methyltransferase SpoU |
| M. avium 104 | MAV_4333 | - | 2e-72 | 83.66% (153) | RNA methyltransferase, TrmH family protein, group 2 |
| M. smegmatis MC2 155 | MSMEG_1636 | - | 4e-70 | 78.57% (154) | RNA methyltransferase, TrmH family protein, group 2 |
| M. thermoresistible (build 8) | TH_2120 | spoU | 3e-63 | 75.82% (153) | PROBABLE tRNA/rRNA METHYLASE SPOU (tRNA/rRNA |
| M. vanbaalenii PYR-1 | Mvan_1533 | - | 2e-70 | 81.82% (154) | tRNA/rRNA methyltransferase (SpoU) |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2852|M.ulcerans_Agy99 --------------MFKLLFYSPRIAPNTGNAIRTAAATGCELHLVEPLG
MMAR_1163|M.marinum_M --------------MFKLLFYSPRIAPNTGNAIRTAAATGCELHLVEPLG
MAV_4333|M.avium_104 --------------MFKLMFVSPRIAPNTGNAIRTAAATGCELHLVEPMG
Mb3401|M.bovis_AF2122/97 --------------MFRLLFVSPRIAPNTGNAIRTCAATGCELHLVEPLG
Rv3366|M.tuberculosis_H37Rv --------------MFRLLFVSPRIAPNTGNAIRTCAATGCELHLVEPLG
MLBr_00419|M.leprae_Br4923 ----------MISKMFRLLFYTPRIPPNTGNAIRTAAVTGCALHLVEPIG
Mflv_4891|M.gilvum_PYR-GCK --------------MFNVLFYSPRIAPNTGNAIRMVAGTGCALHLVEPLG
Mvan_1533|M.vanbaalenii_PYR-1 --------------MFKVMFYSPRIAPNTGNAIRMVAGTGCELHLVEPLG
MSMEG_1636|M.smegmatis_MC2_155 --------------MFRVMFFSPRIAPNTGNAIRMVAATGCELHLVHPLG
TH_2120|M.thermoresistible__bu VCEQSPIAGWDHRLVFRVMFYSPRIPPNTGNAIRMVAATGAELHLVDPL-
MAB_3702|M.abscessus_ATCC_1997 --------------MFHILFHEPRIAPNTGNAIRMVAGTGCELHLVQPL-
:*.::* ***.******** * **. ****.*:
MUL_2852|M.ulcerans_Agy99 FDLSEPKLRRAGLDYHDLASVTVHGSLTQAWDALLPARVFAFTAGASVTF
MMAR_1163|M.marinum_M FDLSEPKLRRAGLDYHDLASVTVHGSLTQAWDALLPARVFAFTAGASVTF
MAV_4333|M.avium_104 FDLSEPKLRRAGLDYHDLASVTVHASLAAAWDALTPARVVAFTSHAPASF
Mb3401|M.bovis_AF2122/97 FDLSEPKLRRAGLDYHDLASVTVHASLAHAWEALSPARVFAFTAQATTLF
Rv3366|M.tuberculosis_H37Rv FDLSEPKLRRAGLDYHDLASVTVHASLAHAWEALSPARVFAFTAQATTLF
MLBr_00419|M.leprae_Br4923 FDLSNPKLRRAGLDYHDLASVTVHTSLSEAWNALSPARVFAFSTLANTLF
Mflv_4891|M.gilvum_PYR-GCK FDLSEPKLRRAGLDYHDLASVTVHADLGAAWSVLMPARVFAFTAHAATSF
Mvan_1533|M.vanbaalenii_PYR-1 FDLSEPKLRRAGLDYHDLASVTVHPDLDSAWAAVTPARVFAFTAHATASF
MSMEG_1636|M.smegmatis_MC2_155 FDLSEPKLRRAGLDYHDLASVTVHDDLPSAWQALMPARVYVFTAHASTPF
TH_2120|M.thermoresistible__bu FDLSEPKLRRAGLDYHDLASVSVHTDLDAAWAALQPRRVFAFTVRAETTY
MAB_3702|M.abscessus_ATCC_1997 FDLSEAKVRRAGLDYHDMASVTVHENLSAAWKTLKPQRVYAFTAHASVTY
****:.*:*********:***:** .* ** .: * ** .*: * . :
MUL_2852|M.ulcerans_Agy99 SDVDFRPGDVLMFGPEPTGLHAATLADAHITQQVRIPMLAGRRSLNLANA
MMAR_1163|M.marinum_M SDVDFRPGDVLMFGPEPTGLDAATLADAHITQQVRIPMLAGRRSLNLANA
MAV_4333|M.avium_104 AELRYRPGDVLMFGPEPTGLDAATLADPHVTTQVRIPMLAGRRSLNLSNA
Mb3401|M.bovis_AF2122/97 TNVGYRAGDVLMFGPEPTGLDEATLADTHITGQVRIPMLAGRRSLNLSNA
Rv3366|M.tuberculosis_H37Rv TNVGYRAGDVLMFGPEPTGLDEATLADTHITGQVRIPMLAGRRSLNLSNA
MLBr_00419|M.leprae_Br4923 TDVSYQAGDVLMFGPEPTGLDAATLADDHITQQVRIPMLSGRRSLNLSNA
Mflv_4891|M.gilvum_PYR-GCK AGVGYEPGDVLMFGPEPTGLDEATLADPHITGQVRIPMLSGRRSLNLSNA
Mvan_1533|M.vanbaalenii_PYR-1 ADVAYRPGDVLMFGPEPTGLDAATLADPHITAQVRIPLLAGRRSLNLSNA
MSMEG_1636|M.smegmatis_MC2_155 TDIAYRPGDVLMFGPEPTGLDEQTLADPHITEQIRIPMLAGRRSLNLSNA
TH_2120|M.thermoresistible__bu VDIHYQPGDVLMFGPEPTGLDAETLADPHITAEVSIPMLPGRRSLNLSNA
MAB_3702|M.abscessus_ATCC_1997 TDIAYQPGDVLLFGPEPTGLDAEVLADPHITEKVRIPLLPGRRSLNLSNA
: :..****:********. .*** *:* :: **:*.*******:**
MUL_2852|M.ulcerans_Agy99 AAVAVYEAWRQHGYPGAL
MMAR_1163|M.marinum_M AAVAVYEAWRQHGYPGAL
MAV_4333|M.avium_104 AAIAVYEAWRQHGYAGAT
Mb3401|M.bovis_AF2122/97 AAVAVYEAWRQHGFAGAV
Rv3366|M.tuberculosis_H37Rv AAVAVYEAWRQHGFAGAV
MLBr_00419|M.leprae_Br4923 AAVAVYEAWRQHGYAGLV
Mflv_4891|M.gilvum_PYR-GCK AAVAVYEAWRQHGFAGAV
Mvan_1533|M.vanbaalenii_PYR-1 AAVAVYEAWRQQGFSGAV
MSMEG_1636|M.smegmatis_MC2_155 AAVAAYEAWRQHGFPGAV
TH_2120|M.thermoresistible__bu AAVAVYEAWRQHGFAGA-
MAB_3702|M.abscessus_ATCC_1997 AAIAAYEAWRQHDFAGSV
**:*.******:.:.*