For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RAARRLRATSKLFYEELGDPSGQPVLLIMGLGAQLPMWPDEFCARLVAAGYRVIRFDHRDTGLSAKMRGM RAPGSVYRRIGRYLLGRPSPVPYTLLDMTEDVAALLDHLSIARAHVVGASLGGMIAQILAADQPQRVASL GIIMSTTGKAFSAPPAWRVIKLAFGQPGPNASAEEKLAFEVRNVAVFNGPNFLPPQSDLRQRVRLLADRC NYPPGMLRQFDAVLGTGSLLGYSKAITAPTVVLHGSADPMVRLRNGRAVAAAIPDARFVVVDGMGHDLPQ PVWRPILETLTENFSRAAPPHV*
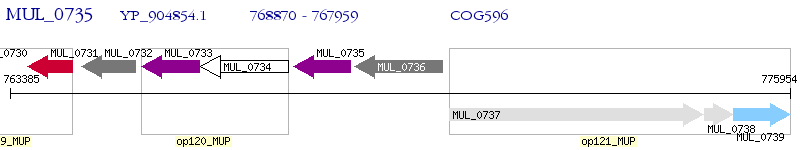
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0735 | lipG2 | - | 100% (303) | lipase/esterase LipG2 |
| M. ulcerans Agy99 | MUL_0733 | lipG1 | 4e-71 | 49.48% (287) | lipase/esterase LipG1 |
| M. ulcerans Agy99 | MUL_0653 | bpoC | 2e-13 | 26.22% (267) | bromoperoxidase BpoC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0665c | lipG | 1e-70 | 47.32% (298) | lipase/esterase LipG |
| M. gilvum PYR-GCK | Mflv_5112 | - | 6e-70 | 48.43% (287) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0646c | lipG | 1e-70 | 47.32% (298) | lipase/esterase LipG |
| M. leprae Br4923 | MLBr_01899 | lipG | 1e-69 | 47.08% (291) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3878 | - | 1e-91 | 55.05% (287) | lipase/esterase LipG |
| M. marinum M | MMAR_0983 | lipG2 | 1e-167 | 97.02% (302) | lipase/esterase LipG2 |
| M. avium 104 | MAV_4515 | - | 2e-67 | 45.48% (299) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_1352 | - | 6e-71 | 47.54% (284) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_1861 | lipG | 1e-72 | 48.59% (284) | PROBABLE LIPASE/ESTERASE LIPG |
| M. vanbaalenii PYR-1 | Mvan_1240 | - | 2e-68 | 47.18% (284) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5112|M.gilvum_PYR-GCK -------MQVRTGTARSGE----LEIHYEDMGDPNDPAVLLIMGLGAQLL
Mvan_1240|M.vanbaalenii_PYR-1 -------MQIRSGTAKSGD----LDIFYEDMGDPNDPAVLLIMGLGAQLL
MSMEG_1352|M.smegmatis_MC2_155 -------MQTRTGLARAGDADHPIDLFYEDMGDPGDPAVLLIMGLGAQML
TH_1861|M.thermoresistible__bu -------MQIRTGFANVDGSPEPVDLYYEDLGDPADPPVLLIMGLGAQLI
Mb0665c|M.bovis_AF2122/97 -------MDIRSGTAVSGD----VKLYYEDMGDLDHPPVLLIMGLGAQML
Rv0646c|M.tuberculosis_H37Rv -------MDIRSGTAVSGD----VKLYYEDMGDLDHPPVLLIMGLGAQML
MAV_4515|M.avium_104 ------MVQVRTGHAISGD----LKLYYEDMGDIDDPPVLLIMGLGAQLL
MLBr_01899|M.leprae_Br4923 -------MEVRSGYATSGD----LKLYYEDMGNVDDPPVLLIMGLGAQLV
MUL_0735|M.ulcerans_Agy99 ---MR----AARRLRAT------SKLFYEELGDPSGQPVLLIMGLGAQLP
MMAR_0983|M.marinum_M ---MRTSSGVARRETTTGD----VELFYEELGDPSGQPVLLIMGLGAQLP
MAB_3878|M.abscessus_ATCC_1997 MVPLSSEVPTRSGIARNGD----VELFYEDLGNPGDPAVVLVMGVAAQLP
.:.**::*: .*:*:**:.**:
Mflv_5112|M.gilvum_PYR-GCK LWRKGFCEKLINQGLRVIRFDNRDVGLSSKLT---GHHAGAPLLPRMARS
Mvan_1240|M.vanbaalenii_PYR-1 LWRNGFCEKLINQGLRVIRFDNRDVGLSSKLH---HHRSGAPLLPRMARS
MSMEG_1352|M.smegmatis_MC2_155 MWRTEFCEKLVDQGLRVIRFDNRDVGLSTKLS---GQRVDSPLALRMARS
TH_1861|M.thermoresistible__bu LWRDGFCRKLVERGLRVIRFDNRDVGLSSKLD---GRRERTPLPWRMLRS
Mb0665c|M.bovis_AF2122/97 LWRTDFCARLVAKGLRVIRYDNRDVGLSTKT---ERHRPGQPLATRLVRS
Rv0646c|M.tuberculosis_H37Rv LWRTDFCARLVAKGLRVIRYDNRDVGLSTKT---ERHRPGQPLATRLVRS
MAV_4515|M.avium_104 LWRTAFCEKLVGRGLRVIRYDNRDVGLSSKT---EHRSSGQPLVTRLLRS
MLBr_01899|M.leprae_Br4923 LWRTAFCEKLVAQGLRVVRYDNRDVGLSSRTDSTEQPCPSQPLTARLIRF
MUL_0735|M.ulcerans_Agy99 MWPDEFCARLVAAGYRVIRFDHRDTGLSAKMR---GMRAPGSVYRRIGRY
MMAR_0983|M.marinum_M MWPDEFCARLVTAGYRVIRFDHRDTGLSAKMR---GMRAPGSVYRRIGRY
MAB_3878|M.abscessus_ATCC_1997 MWPDGFCRQLLDQGYRVIRFDNRDCGLSTKLD---GQKAPGSVQRRVVRY
:* ** :*: * **:*:*:** ***:: .: *: *
Mflv_5112|M.gilvum_PYR-GCK LAGLPSPAAYTLEDMADDAAALLDHLDIDRAHVVGGSMGGMIAQVFAARH
Mvan_1240|M.vanbaalenii_PYR-1 FVGRPSPAAYTLEDMADDAAALLDHLGIDTAHVVGGSMGGMIAQVFAARH
MSMEG_1352|M.smegmatis_MC2_155 FLGKRSPAVYTLEDMADDAAALLDHLEIDRAHIVGGSMGGMIAQVFASQH
TH_1861|M.thermoresistible__bu FAGLPSPAVYTLEDMADDAAGLLDHLQIDAAHIVGASMGGMVAQVFAGRH
Mb0665c|M.bovis_AF2122/97 WLGLPSQAAYTLEDMAADAAALLDHLDVKHAHVVGASMGGMIAQIFAARF
Rv0646c|M.tuberculosis_H37Rv WLGLPSQAAYTLEDMAADAAALLDHLDVKHAHVVGASMGGMIAQIFAARF
MAV_4515|M.avium_104 WLGLPSQSAYRLEDMADDAAAVLDHLGIDDAHIVGASMGGMIAQIFAARF
MLBr_01899|M.leprae_Br4923 WLGQRNACAYTLEDMTDDAVALLDHLSIERAHIVGASMGGMIAQIFAARF
MUL_0735|M.ulcerans_Agy99 LLGRPSPVPYTLLDMTEDVAALLDHLSIARAHVVGASLGGMIAQILAADQ
MMAR_0983|M.marinum_M LLGRPSPVPYTLLDMTEDVAALLDHLSIARAHVVGASLGGMIAQILAADR
MAB_3878|M.abscessus_ATCC_1997 ALGMGSKVPYTLIDMAEDVRSLVDHLGLEKVHIAGASMGGMIVQVFAGTY
* . * * **: *. .::*** : .*:.*.*:***:.*::*.
Mflv_5112|M.gilvum_PYR-GCK HPRTKTLGVIFSSNNQAALPPPGPRQLAAILQ-RPKGSSRDEIVDNAVRV
Mvan_1240|M.vanbaalenii_PYR-1 RARTGALGVIFSSNNQAALPPPGPRQLAAILQ-RPKGSDREAIIENAVRV
MSMEG_1352|M.smegmatis_MC2_155 AARTETLGVIFSSNNQALLPPPGIRQLLSLITGPPADASRDAVLDNAVRL
TH_1861|M.thermoresistible__bu AHRTRSLGVIFSSNNRPFLPPPGPRQLLSIITGPAPDAPRDEVIENAVKV
Mb0665c|M.bovis_AF2122/97 AQRTKTLAVIFSSNNHRFLPPPAPRALLALLTGPPPDSPRDVIVDNAVRV
Rv0646c|M.tuberculosis_H37Rv AQRTKTLAVIFSSNNHRFLPPPAPRALLALLTGPPPDSPRDVIVDNAVRV
MAV_4515|M.avium_104 RQRTKTLAVIFSSNNSALLPPPAPRALLALLKGPPPDSPREVIIDNAVRV
MLBr_01899|M.leprae_Br4923 PTRTRSLAVFFSSNNRPFLPPPAPRALLALLTGPPTGSRRDVVVDNVVRV
MUL_0735|M.ulcerans_Agy99 PQRVASLGIIMSTTGKAFSAPPAWRVIKLAFGQPGPNASAEEKLAFEVRN
MMAR_0983|M.marinum_M PQRVASLGIIMSTTGKAFSAPPAWRVIKLAFGQPGPNASAEEKLAFEVRN
MAB_3878|M.abscessus_ATCC_1997 PERVHSVGIIYSATGRPFSRLPSWELIKTALNAPGKNATAEEWLEFEVNN
*. ::.:: *:.. *. . : : .: : : *.
Mflv_5112|M.gilvum_PYR-GCK SRIIGSPGYPAPLDRLRTDAIEGFDRSYYPAGVGRHFAAILGSGSLRRYD
Mvan_1240|M.vanbaalenii_PYR-1 SRIIGSPGYPAAEERLRADAIEGYDRSYYPAGVGRHFAAILGSGSLRRYD
MSMEG_1352|M.smegmatis_MC2_155 SRLNGSPAYPRSEEEIRAEAAELYDRAYYPAGIARHFAAILGSGSLRHHN
TH_1861|M.thermoresistible__bu GRLNGSPAYPVPDEQLRADAATAFDRSYHPVGVARQFDAILGSGSLRRYN
Mb0665c|M.bovis_AF2122/97 SKIIGSPAYPIPEDQVRAEAAESYDRNFHPWGIAQQFSAILGSGSLLRYD
Rv0646c|M.tuberculosis_H37Rv SKIIGSPAYPIPEDQVRAEAAESYDRNFHPWGIAQQFSAILGSGSLLRYD
MAV_4515|M.avium_104 GRIIGSTRYRVPEEQARAEAAEGYDRNYYPQGVARHFSAVLGSGSLRRYN
MLBr_01899|M.leprae_Br4923 TKITGSPLYRMPEEQVRTNAAEIYDRSFYPLGVSRQFSAILGSGSLLHYN
MUL_0735|M.ulcerans_Agy99 VAVFNGPNFLPPQSDLRQRVRLLADRCNYPPGMLRQFDAVLGTGSLLGYS
MMAR_0983|M.marinum_M VAVFNGPNFLPPQSDLRQRVRLLADRCNYPPGMLRQFDAVLGTGSLLGYS
MAB_3878|M.abscessus_ATCC_1997 GIVYNGPDNLPSREQLRQRILDHRARSDYKIGTVRQFDAILGTGSLLRFT
: ... . . * * : * ::* *:**:*** .
Mflv_5112|M.gilvum_PYR-GCK RQITAPTVVIHGRADKLMRPSGGRAIARAIPGARLVLFDGMGHDLPEPLW
Mvan_1240|M.vanbaalenii_PYR-1 RQITAPTVVIHGTADKLMRPSGGRAIARAIPRARLVLFDGMGHELPEPLW
MSMEG_1352|M.smegmatis_MC2_155 ERVTAPTVVIHGRADRLMPPSGGRAIANAIRGARMVLIDGMGHELPEPVW
TH_1861|M.thermoresistible__bu RRTTSPTVVIHGRADKLMRPTGGRAVARAIPGARLTLFDGMGHDLPEPLW
Mb0665c|M.bovis_AF2122/97 RRIVAPTVVIHGRADKLMRPFGGRAVARAINGARLVLIDGMGHDLPRQLW
Rv0646c|M.tuberculosis_H37Rv RRIVAPTVVIHGRADKLMRPFGGRAVARAINGARLVLIDGMGHDLPRQLW
MAV_4515|M.avium_104 RRTTAPTVVIHGRADKLMRPFGGRAVAGAIDGARLVLFDGMGHDLPQQLW
MLBr_01899|M.leprae_Br4923 QRIIAPTVVIHGRADKLVRPSSGRAVARAITGARLVLFDGMGHDLPQQLW
MUL_0735|M.ulcerans_Agy99 KAITAPTVVLHGSADPMVRLRNGRAVAAAIPDARFVVVDGMGHDLPQPVW
MMAR_0983|M.marinum_M KAITAPTVVLHGSADPMVRLRNGRAVAAAIPDARFVVVDGMGHDLPQPVW
MAB_3878|M.abscessus_ATCC_1997 RAVVAPAVVIHGRNDPLVPYQNGRVVAKNMRNARFALIERMGHDLPEPVW
. :*:**:** * :: .**.:* : **:.:.: ***:**. :*
Mflv_5112|M.gilvum_PYR-GCK DDIVGELKTTFAEAP----
Mvan_1240|M.vanbaalenii_PYR-1 DDIVGELKTTFAEIS----
MSMEG_1352|M.smegmatis_MC2_155 DEVIGELKTNFAVVG----
TH_1861|M.thermoresistible__bu DEIVAELDKNITAGR----
Mb0665c|M.bovis_AF2122/97 DRVIGELTRNFSEAG----
Rv0646c|M.tuberculosis_H37Rv DRVIGELTRNFSEAG----
MAV_4515|M.avium_104 DQVIGVLANNFAKAS----
MLBr_01899|M.leprae_Br4923 DQAVGVLMSNFAKAS----
MUL_0735|M.ulcerans_Agy99 RPILETLTENFSRAAPPHV
MMAR_0983|M.marinum_M RPILETLTENFSRAAPPHV
MAB_3878|M.abscessus_ATCC_1997 KPVTSELLTTFECAVQD--
* .: