For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ETTESRFVNLGRDGGVATVELIDSGPLNIVGTAAITALTNAFRSINQDSGVRVVVLRAAGEKAFIGGADI KEMVTLDRVSAQAFIRRLAGLCESIRQCPVPVIARIAGWCLGGGLEIAACCDLRIAESGARFGMPEVAVG IPSVIHSALLPALVGASRTTWLLLTGETIDADTAASWALVHEVVAPAALDLRIAALATKLAGFGPQALRQ QKRLLAKWPDMTVPAAIEDSIEQFGLAFQTGEPQQHMQAFLSRKGRKAGSGPQAPEE*
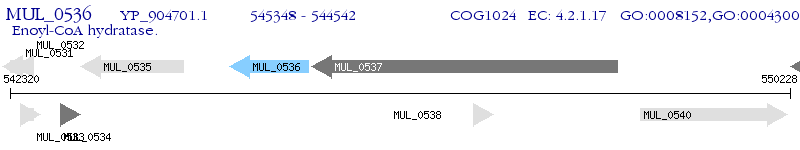
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0536 | echA8_3 | - | 100% (268) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 3e-26 | 31.15% (244) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_2758 | echA8_5 | 9e-22 | 31.97% (244) | enoyl-CoA hydratase, EchA8_5 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3408 | echA18 | 8e-28 | 32.79% (247) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 7e-27 | 31.97% (244) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 8e-25 | 30.33% (244) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-24 | 31.44% (229) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_0905 | - | 1e-23 | 35.08% (248) | putative enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_4952 | echA8_3 | 1e-149 | 99.25% (268) | enoyl-CoA dehydratase, EchA8_3 |
| M. avium 104 | MAV_1195 | - | 1e-23 | 30.33% (244) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5277 | - | 2e-27 | 32.79% (244) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_4102 | - | 1e-25 | 31.17% (247) | PUTATIVE short chain enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4681 | - | 1e-25 | 31.97% (244) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0536|M.ulcerans_Agy99 --------------------------------METTESRFVNLGRDGGVA
MMAR_4952|M.marinum_M --------------------------------METTESRFVNLGRDGGVA
Rv1070c|M.tuberculosis_H37Rv -----------------------------------MTYETILVERDQRVG
MLBr_02402|M.leprae_Br4923 -----------------------------------MKYDTILVDGYQRVG
MAV_1195|M.avium_104 -----------------------------MTDSTDRTFETILVEREERVG
Mflv_2036|M.gilvum_PYR-GCK -----------------------------------MTFETILVDRDDRVA
Mvan_4681|M.vanbaalenii_PYR-1 --------------------------------MGSQNFETIIVERDDRVA
MSMEG_5277|M.smegmatis_MC2_155 ----------------------------------MSDFETILVTRTDRVA
TH_4102|M.thermoresistible__bu --------------------------------MSAKNYETILVTREDRVG
Mb3408|M.bovis_AF2122/97 MRRRAMTKMDEASNPCGGDIEAEMCQLMREQPPAEGVVDRVALQRHRNVA
MAB_0905|M.abscessus_ATCC_1997 -------------------------------MTQTAPAAVAYSVNHAGVA
*.
MUL_0536|M.ulcerans_Agy99 TVELIDSGPLNIVGTAAITALTNAFRSINQDSGVRVVVLRAAGEKAFIGG
MMAR_4952|M.marinum_M TVELIDSGPLNIVGTAAITALTNAFRSINQDSGVRVVVLRAAGEKAFIGG
Rv1070c|M.tuberculosis_H37Rv IITLNRPQALNALNSQVMNEVTSAATELDDDPDIGAIIITGS-AKAFAAG
MLBr_02402|M.leprae_Br4923 IITLNRPQALNALNSQMMNEITNAAKELDIDPDVGAILITGS-PKVFAAG
MAV_1195|M.avium_104 IITLNRPKALNALNTQVMNEVTTAATDFDADPGIGAIIITGS-AKAFAAG
Mflv_2036|M.gilvum_PYR-GCK TITLNRPKALNALNTQVMNEVTTAAAELDAEPGVGAIIVTGS-ERAFAAG
Mvan_4681|M.vanbaalenii_PYR-1 TITLNRPKALNALNSQVMDEVTTAAAELDADAGVGAIILTGS-EKAFAAG
MSMEG_5277|M.smegmatis_MC2_155 TIQLNRPKALNALNSQVMTEVTAAAAELDKDPGVGAIIVTGN-EKAFAAG
TH_4102|M.thermoresistible__bu IITLNRPKALNALNSQVMTEVTDAAAEFDSDPGIGAIIVTGN-EKAFAAG
Mb3408|M.bovis_AF2122/97 LITLSHPQAQNALNLASWRRLKRLLDDLAGESGLRAVVLRGAGDKAFAAG
MAB_0905|M.abscessus_ATCC_1997 AIVLDRPEASNALDRTMKTELLQALLAAGGDPAVRAVVMSAA-GKNFCVG
: * . . * :. : :. : .::: . : * *
MUL_0536|M.ulcerans_Agy99 ADIKEMVTLDR----VSAQAFIRRLAGLCESIRQCPVPVIARIAGWCLGG
MMAR_4952|M.marinum_M ADIKEMVTLDR----ISAQAFIRRLAGLCESIRQCPVPVIARMAGWCLGG
Rv1070c|M.tuberculosis_H37Rv ADIKEMADLTFAD--AFTADFFATWG----KLAAVRTPTIAAVAGYALGG
MLBr_02402|M.leprae_Br4923 ADIKEMASLTFTD--AFDADFFSAWG----KLAAVRTPMIAAVAGYALGG
MAV_1195|M.avium_104 ADIKEMAELTFAD--AYGADFFAPWG----KLAALRTPTIAAVAGHALGG
Mflv_2036|M.gilvum_PYR-GCK ADIKEMASLSFAD--VFSSDFFAAWG----KFAATRTPTIAAVAGYALGG
Mvan_4681|M.vanbaalenii_PYR-1 ADIKEMAGLSFAD--VFSSDFFAGWG----KFAATRTPTIAAVAGYALGG
MSMEG_5277|M.smegmatis_MC2_155 ADIKEMADLSFAE--VFEADFFELWS----KFAATRTPTIAAVAGYALGG
TH_4102|M.thermoresistible__bu ADIKEMADLEFAD--VFAADFFAPWS----KFAATRTPTIAAVAGYALGG
Mb3408|M.bovis_AF2122/97 ADIKEFPNTRMSA--ADAAEYNESLAVCLRALTTMPIPVIAAVRGLAVGG
MAB_0905|M.abscessus_ATCC_1997 QDLAEHVEALRDDPAHAMDTVREHYNPVLEALDAIKVPVVVAINGACVGA
*: * : * :. : * .:*.
MUL_0536|M.ulcerans_Agy99 GLEIAACCDLRIAESGARFGMPEVAVGIPS-VIHSALLPALVGASRTTWL
MMAR_4952|M.marinum_M GLEIAACCDLRIAESGARFGMPEVAVGIPS-VIHSALLPALVGASRTTWL
Rv1070c|M.tuberculosis_H37Rv GCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
MLBr_02402|M.leprae_Br4923 GCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
MAV_1195|M.avium_104 GCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
Mflv_2036|M.gilvum_PYR-GCK GCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
Mvan_4681|M.vanbaalenii_PYR-1 GCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
MSMEG_5277|M.smegmatis_MC2_155 GCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
TH_4102|M.thermoresistible__bu GCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
Mb3408|M.bovis_AF2122/97 GCELATACDVCIATDDARFGIPLGKLGVTTGFTEADTVARLIGPAALKYL
MAB_0905|M.abscessus_ATCC_1997 GLGLALGADIRIAGQRAKFGTAFTGIGLAADSALSASLPRLIGASRATAM
* :* .*: ** . *:** . :*: . : :. :* : :
MUL_0536|M.ulcerans_Agy99 LLTGETIDADTAASWALVHEVVAPAALDLRIAALATKLAGFGPQALRQQK
MMAR_4952|M.marinum_M LLTGETIDADTAASWALVHEVVAPAALDLRIAALATKLAGFGPQALRQQK
Rv1070c|M.tuberculosis_H37Rv ILTGRTMDAAEAERSGLVSRVVPADDLLTEARATATTISQMSASAARMAK
MLBr_02402|M.leprae_Br4923 ILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAK
MAV_1195|M.avium_104 ILTGRTIDAAEAERSGLVSRVVPADDLLTEAKKVATTISQMSRSAARMAK
Mflv_2036|M.gilvum_PYR-GCK ILTGRNMDAEEAERAGLVSRVVPADSLLSEAKAVAQTIAGMSLSASRMAK
Mvan_4681|M.vanbaalenii_PYR-1 ILTGRNMGAEEAERAGLVSRVVPADTLLEEARSVAKTIAGMSLSASRMAK
MSMEG_5277|M.smegmatis_MC2_155 ILTGRTIDAAEAERAGLVSRLVPADTLIDEALAVAQTIAGMSLSASRMAK
TH_4102|M.thermoresistible__bu ILTGRTIDAEEAERSGLVSRVVPADKLLDEAKETAATIAAMSLSAARMAK
Mb3408|M.bovis_AF2122/97 LFSGELIGIEEAARWGLVQKVVAPQDLAAATAKLVGQVCRQSAVTMRAAK
MAB_0905|M.abscessus_ATCC_1997 FLLGDTIDAPTAHTWGLVHEVVDEGSPADVANSVAGRLAGGPTAAFSEVK
:: * :. * .** .:* . :. : *
MUL_0536|M.ulcerans_Agy99 RLLAKWPDMTVPAAIEDSIEQFGLAFQTGEPQQHMQAFLSRKGRKAGSGP
MMAR_4952|M.marinum_M RLLAKWPDMTVPAAIEDSIEQFGLAFQTGEPQQHMQAFLSRKGRKAGSGP
Rv1070c|M.tuberculosis_H37Rv EAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR-
MLBr_02402|M.leprae_Br4923 EAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR-
MAV_1195|M.avium_104 EAVNRAFETTLAEGLLYERRLFHSTFATADQSEGMAAFVEKRAPNFTHR-
Mflv_2036|M.gilvum_PYR-GCK EAVDRAFETTLSEGLLYERRLFHSAFATDDQTEGMNAFSEKRSPNFTHR-
Mvan_4681|M.vanbaalenii_PYR-1 EAVDRAFETTLTEGLLYERRLFHSAFATDDQTEGMNAFTEKRSPNFTHR-
MSMEG_5277|M.smegmatis_MC2_155 EAVNRAFETSLTEGLLYERRLFHSAFATADQKEGMAAFAEKRAANFTHR-
TH_4102|M.thermoresistible__bu EAVNRAFETSLAEGLLFERRVFHSAFATADQSEGMAAFTEKRPPKFTHR-
Mb3408|M.bovis_AF2122/97 VVANMHGRALTGADTDALIRFGVEAYEGADLREGVAAFSQGRPPKFDD--
MAB_0905|M.abscessus_ATCC_1997 ELLRRNAVAPLGDVLEREASAQQRLGASRDHSAAVEAFLAKDKPVFVGR-
: : **
MUL_0536|M.ulcerans_Agy99 QAPEE
MMAR_4952|M.marinum_M QAPEE
Rv1070c|M.tuberculosis_H37Rv -----
MLBr_02402|M.leprae_Br4923 -----
MAV_1195|M.avium_104 -----
Mflv_2036|M.gilvum_PYR-GCK -----
Mvan_4681|M.vanbaalenii_PYR-1 -----
MSMEG_5277|M.smegmatis_MC2_155 -----
TH_4102|M.thermoresistible__bu -----
Mb3408|M.bovis_AF2122/97 -----
MAB_0905|M.abscessus_ATCC_1997 -----