For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVDLEFDGLDTGLAVLTIDRPHARNAIGLETMAQLQDALDAAAGAQALVIKGAGDRAFVSGGDLKELSAL RTLEDASAMAKRMRSICDQIATFPAPVIAALNGHAFGGGAEVAVAADIRVAADDIKIAFNQVLLEIMPAW GGAERLAAVVGKSKALLLAGSGIALDAVEAERIGLVDLVFPRASFDSTDGGWRSLARSLARRPAAEVKRV INGTSPDEAIASFARLWVEDAHWQAAEQVMNRTPNRGPGAEGAT
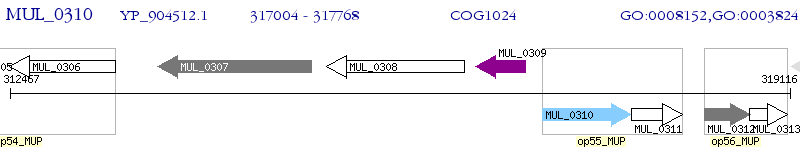
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0310 | echA8_2 | - | 100% (254) | enoyl-CoA hydratase, EchA8_2 |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 2e-26 | 33.65% (208) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_1899 | echA17 | 7e-24 | 34.02% (244) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 4e-25 | 36.00% (175) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2522 | - | 2e-89 | 69.83% (242) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 5e-25 | 36.00% (175) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_01724 | - | 2e-24 | 34.96% (226) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 3e-23 | 35.29% (170) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4691 | echA8_2 | 1e-138 | 98.82% (254) | enoyl-CoA hydratase, EchA8_2 |
| M. avium 104 | MAV_0967 | - | 1e-111 | 81.50% (254) | enoyl-CoA hydratase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_4775 | - | 1e-91 | 70.25% (242) | enoyl-CoA hydratase/isomerase family protein |
| M. thermoresistible (build 8) | TH_3655 | - | 2e-80 | 70.26% (232) | PUTATIVE short chain enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4136 | - | 9e-91 | 71.49% (242) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0310|M.ulcerans_Agy99 -----------------MVDLEFDG--LDTGLAVLTIDRPHARNAIGLET
MMAR_4691|M.marinum_M -----------------MVDLEFDE--LDTGLAVLTIDRPHARNAIGLET
MAV_0967|M.avium_104 -----------------MVDLEID-----DGLAVLTIDRPHARNAIALDT
Mflv_2522|M.gilvum_PYR-GCK -----------------MVDLEID-----DGLAIITIDRPHARNAISLET
Mvan_4136|M.vanbaalenii_PYR-1 MVFSSCENANTVNGECPMVDLELD-----DGLAVITIDRPHARNAISLET
MSMEG_4775|M.smegmatis_MC2_155 -------------MENDLVDLELD-----GELAVITIDRPHARNAISLDT
TH_3655|M.thermoresistible__bu -----------------MVDLELT-----DGLAVITIDRPQARNAIAPAT
Mb1099c|M.bovis_AF2122/97 ------------MT---YETILVE---RDQRVGIITLNRPQALNALNSQV
Rv1070c|M.tuberculosis_H37Rv ------------MT---YETILVE---RDQRVGIITLNRPQALNALNSQV
MAB_1185c|M.abscessus_ATCC_199 ------------MTSHNFETILTE---RIDRVAVITLNRPKALNALNSQV
MLBr_01724|M.leprae_Br4923 ------------MS--EFVNIVVSDGSQDAGLAILLLSRP-PTNAMTRQF
: :.:: :.** . **:
MUL_0310|M.ulcerans_Agy99 MAQLQDALDAAAG---AQALVIKGAGDRAFVSGGDLKELSALRTLEDASA
MMAR_4691|M.marinum_M MAQLQDALDAAAG---AQALVIKGAGDRAFVSGGDLKELSALRTLEDASA
MAV_0967|M.avium_104 MDQLEKALDAAAG---AQALVITGAGDRAFVSGGDLKELSALRTEEDAAA
Mflv_2522|M.gilvum_PYR-GCK MDQLDTALDGAHG---ARALALTGAGDKAFVSGGDLKELSALRTVEQAAS
Mvan_4136|M.vanbaalenii_PYR-1 MEQLDKALDGAEG---AAALALTGAGDKAFVSGGDLKELSALRTEEQAAA
MSMEG_4775|M.smegmatis_MC2_155 MDELDKALDGAAG---SRALAITGAGDRAFVSGGDLKELAALRTEREAGA
TH_3655|M.thermoresistible__bu MEQLEKALDSLAGNPDAQALVIRGAGDRAFVSGGDLKELAALRTELEASA
Mb1099c|M.bovis_AF2122/97 MNEVTSAATELDDDPDIGAIIITGS-AKAFAAGADIKEMADL-TFADAFT
Rv1070c|M.tuberculosis_H37Rv MNEVTSAATELDDDPDIGAIIITGS-AKAFAAGADIKEMADL-TFADAFT
MAB_1185c|M.abscessus_ATCC_199 MNEVTTAAAEFDADHGIGAIIITGS-EKAFAAGADIKEMSEQ-SFSDMFG
MLBr_01724|M.leprae_Br4923 YREIAMAADELGRRDDVAAVILFGG-HDIFSAGDDMQELRTL-TAVEAGV
:: * *: : *. * :* *::*: : :
MUL_0310|M.ulcerans_Agy99 MAKRMRSICDQIATFPAPVIAALNGHAFGGGAEVAVAADIRVAADDIKIA
MMAR_4691|M.marinum_M MAKRMRSICDQIAAFPAPVIAALNGHAFGGGAEVAVAADIRVAADDIKIA
MAV_0967|M.avium_104 MARRMRAICDQLADFPAPVVAALNGHAFGGGAEVAVAADIRVAADDIKIA
Mflv_2522|M.gilvum_PYR-GCK MSFRMRSICDRIAGFPAPVVAALNGHALGGGAEFAVAADIRLAADDIKIG
Mvan_4136|M.vanbaalenii_PYR-1 MAFRMRSICDRIAGFPGPVLAALNGHALGGGAEFAVSADIRVAADDIKIG
MSMEG_4775|M.smegmatis_MC2_155 MAWRMRSICDRIAGFPGPVIAALNGHALGGGAEVAVSADIRIAADDVKIG
TH_3655|M.thermoresistible__bu MAWRMRSICDRLARFPGPVIAALNGHALGGGAEVAVAADIRIAADDIRIG
Mb1099c|M.bovis_AF2122/97 ADFFA--TWGKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFG
Rv1070c|M.tuberculosis_H37Rv ADFFA--TWGKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFG
MAB_1185c|M.abscessus_ATCC_199 SDFFS--AWGKLGAVRTPTIAAVSGYALGGGCELAMMCDVIIAAENATFG
MLBr_01724|M.leprae_Br4923 AAEVRRQAVDAVATIPKPTVAAITGYALGAGLTLALAADWRVSGDNVKFG
. :. . *.:**: *:*:*.* .*: .* ::.: :.
MUL_0310|M.ulcerans_Agy99 FNQVLLEIMPAWGGAERLAAVVGKSKALLLAGSGIALDAVEAERIGLVDL
MMAR_4691|M.marinum_M FNQVLLEIMPAWGGAERLAAVVGKSKALLLAGSGIALDAVEAERIGLVDL
MAV_0967|M.avium_104 FNQVALEIMPAWGGAERLAALVGQSRALLLAGSGTALDAVEAERIGLVDR
Mflv_2522|M.gilvum_PYR-GCK FNQVALEIMPAWGGAERLVELVGYSRALLLAGTGRILAATDAERLGLIDQ
Mvan_4136|M.vanbaalenii_PYR-1 FNQVALAIMPAWGGAERLVKLVGYSRALLLAGTGRILDAADAERIGLIDQ
MSMEG_4775|M.smegmatis_MC2_155 FNQVALEIMPAWGGAERLVTLVGRSRALLLAGTGRVLDAAEAERVGLVDQ
TH_3655|M.thermoresistible__bu FNQVALAIMPAWGGAERLAELVGRSKALLLAGTGTVLTAAEAERVGLVDK
Mb1099c|M.bovis_AF2122/97 QPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSR
Rv1070c|M.tuberculosis_H37Rv QPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSR
MAB_1185c|M.abscessus_ATCC_199 QPEIKLGVLPGMGGSQRLTRAIGKAKAMDMILTGRNMDAAEAERSGLVSR
MLBr_01724|M.leprae_Br4923 ATEILADLVPDGDGLARLTRVAGVSRAKDLVFSGRFFDAAEALALGLIDD
:: ::* .* **. * ::* : :* : *.:* **:.
MUL_0310|M.ulcerans_Agy99 VFPRASFDSTDGGWRSLARSLARRPAAEVKR--------------VINGT
MMAR_4691|M.marinum_M VFPRASFDSTDGGWRSLARSLARRPAAEVKR--------------VINGT
MAV_0967|M.avium_104 VLPRDVFDSAEQGWRSIARSLARRPAAEIKR--------------VIRGV
Mflv_2522|M.gilvum_PYR-GCK VIPRASFD---EQWRLTARLLASVPAGEVKR--------------VMRGV
Mvan_4136|M.vanbaalenii_PYR-1 VIPRASFD---EQWRLIARSLASAPAGEVKR--------------VMRGV
MSMEG_4775|M.smegmatis_MC2_155 VLPRAEFA---DGWRALAHTLAGRPAGEIKR--------------VMGGV
TH_3655|M.thermoresistible__bu VVPRASFD---TEWRAIARSLATRPAAEIKRGARGGRRQPDEPDEVAVGF
Mb1099c|M.bovis_AF2122/97 VVPADDLLTEARATATTISQMSASAARMAKEAVN-----------RAFES
Rv1070c|M.tuberculosis_H37Rv VVPADDLLTEARATATTISQMSASAARMAKEAVN-----------RAFES
MAB_1185c|M.abscessus_ATCC_199 VVATESLLDEAKAVAKTISEMSLSASMMAKEAVN-----------RAFES
MLBr_01724|M.leprae_Br4923 MVAPDDVYDIAAAWARRFLDGLPHALAAAKAGIN-----------EVFDL
:.. . . *
MUL_0310|M.ulcerans_Agy99 SPDEAIASFARLWVEDAHWQAAEQVMNRTPNRGPGAEGAT----------
MMAR_4691|M.marinum_M SPDEAIASFARLWVEDAHWQAAERVMNRTPNRGPGAEGAT----------
MAV_0967|M.avium_104 SPEEAIASFARLWVADPHWQAAERVMARNTKPRPAAGGAS----------
Mflv_2522|M.gilvum_PYR-GCK PTTEAVAAFARLWVSDEHWAAADKVMKKGK--------------------
Mvan_4136|M.vanbaalenii_PYR-1 PTTEAVAAFARLWASDEHWAAADKVMKKGK--------------------
MSMEG_4775|M.smegmatis_MC2_155 PATEAVAAFARLWVAEEHWAAADKVMNKKK--------------------
TH_3655|M.thermoresistible__bu SPGSRIARSDRRPHNRSAGSAREVPSLRAQLRGQVPTGEQPRGRGHPAPD
Mb1099c|M.bovis_AF2122/97 SLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR----------
Rv1070c|M.tuberculosis_H37Rv SLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR----------
MAB_1185c|M.abscessus_ATCC_199 SLAEGLLFERRIFHSAFGTADQSEGMAAFVEKRPANFIHR----------
MLBr_01724|M.leprae_Br4923 EPAERIAAERRRYVEVFAAAQPSLLLGGCD--------------------
. : * .
MUL_0310|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4691|M.marinum_M --------------------------------------------------
MAV_0967|M.avium_104 --------------------------------------------------
Mflv_2522|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4775|M.smegmatis_MC2_155 --------------------------------------------------
TH_3655|M.thermoresistible__bu PPPVAGAYQLIVVVTDRVDPVLPVVRAQHQHRVHIRARGGPVAVDVDDDV
Mb1099c|M.bovis_AF2122/97 --------------------------------------------------
Rv1070c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01724|M.leprae_Br4923 --------------------------------------------------
MUL_0310|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4691|M.marinum_M --------------------------------------------------
MAV_0967|M.avium_104 --------------------------------------------------
Mflv_2522|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4775|M.smegmatis_MC2_155 --------------------------------------------------
TH_3655|M.thermoresistible__bu AHAVVGRSHRSVHLRHHLGEGVRIGEGAELVVVDVDPVGPAAAEQRPVLL
Mb1099c|M.bovis_AF2122/97 --------------------------------------------------
Rv1070c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01724|M.leprae_Br4923 --------------------------------------------------
MUL_0310|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4691|M.marinum_M --------------------------------------------------
MAV_0967|M.avium_104 --------------------------------------------------
Mflv_2522|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4775|M.smegmatis_MC2_155 --------------------------------------------------
TH_3655|M.thermoresistible__bu VDTRCVADQYVGDLRPVGVHLSHGNRRGGSRRDHHRPCNRFSLDENVVSG
Mb1099c|M.bovis_AF2122/97 --------------------------------------------------
Rv1070c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01724|M.leprae_Br4923 --------------------------------------------------
MUL_0310|M.ulcerans_Agy99 -------------------------------
MMAR_4691|M.marinum_M -------------------------------
MAV_0967|M.avium_104 -------------------------------
Mflv_2522|M.gilvum_PYR-GCK -------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 -------------------------------
MSMEG_4775|M.smegmatis_MC2_155 -------------------------------
TH_3655|M.thermoresistible__bu YCHRRRHRARPPGPGRRGPAGGVNAVVQTPG
Mb1099c|M.bovis_AF2122/97 -------------------------------
Rv1070c|M.tuberculosis_H37Rv -------------------------------
MAB_1185c|M.abscessus_ATCC_199 -------------------------------
MLBr_01724|M.leprae_Br4923 -------------------------------