For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEEAFIYEAIRTPRGKQKYGSLNEVKPISLVVGLIEELRKRNPDLDENLISDVILGCVSPVGDQGGDIAR AAVLASGMPVTSGGVQLNRFCASGLEAVNMAAQKVRSGWDDLVLAGGVESMSRVPMGADGGAMGLDPVTN YDVMFVPQGIGADLIATIEGFSRDDVDAYALRSQEKAAEAWSGGYFAKSVVPVRDQNGLLILDHDEHMRP ETTKEGLGKLKSAFEGLAAMGGFDDVALQKYHWVEKINHVHTGGNSSGIVDGAAIVMVGSEKAGAAINAT PRARIVATATSGADPVIMLTGPTPAARKVLDRAGLTVDDIDLFELNEAFASVVLKFQKDLNIPDEKLNVN GGAIAMGHPLGATGAMILGTMVDELERRNARRALVTLCIGGGMGAATIIERV*
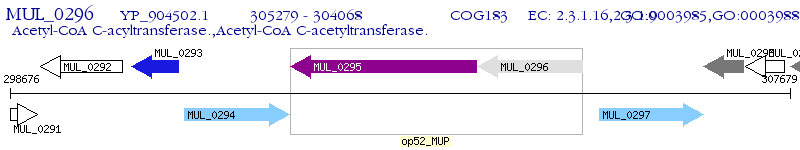
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0296 | fadA | - | 100% (403) | acetyl-CoA acetyltransferase |
| M. ulcerans Agy99 | MUL_2563 | fadA6_4 | 1e-79 | 44.47% (407) | acetyl-CoA acetyltransferase FadA6_4 |
| M. ulcerans Agy99 | MUL_0971 | fadA6_3 | 6e-71 | 40.30% (402) | acetyl-CoA acetyltransferase FadA6_3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0882 | fadA | 0.0 | 93.55% (403) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_1691 | - | 0.0 | 85.36% (403) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv0859 | fadA | 0.0 | 93.30% (403) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | MLBr_02162 | fadA | 0.0 | 92.31% (403) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_0850 | - | 0.0 | 84.50% (400) | acetyl-CoA acetyltransferase |
| M. marinum M | MMAR_4677 | fadA | 0.0 | 99.26% (403) | acyl-CoA thiolase FadA |
| M. avium 104 | MAV_0980 | - | 0.0 | 87.34% (403) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_5721 | - | 0.0 | 84.37% (403) | acetyl-CoA acetyltransferase |
| M. thermoresistible (build 8) | TH_0255 | - | 2e-75 | 43.39% (401) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5067 | - | 0.0 | 84.12% (403) | acetyl-CoA acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0296|M.ulcerans_Agy99 --------------MSEEAFIYEAIRTPRGKQKYGSLNEVKPISLVVGLI
MMAR_4677|M.marinum_M MTDLNPNFCEQEVDMSEEAFIYEAIRTPRGKQKNGSLNEVKPISLVVGLI
Mb0882|M.bovis_AF2122/97 --------------MSEEAFIYEAIRTPRGKQKNGSLHEVKPLSLVVGLI
Rv0859|M.tuberculosis_H37Rv --------------MSEEAFIYEAIRTPRGKQKNGSLHEVKPLSLVVGLI
MLBr_02162|M.leprae_Br4923 --------------MSEEAFIYEAIRTPRGKQKNGSLHEVKPVSLVVGLV
MAV_0980|M.avium_104 --------------MSEEAFIYEAIRTPRGKQKNGSLTEVKPLNLVVGLV
Mflv_1691|M.gilvum_PYR-GCK --------------MSEEAFIYEAIRTPRGKQRGGALNEIKPVNLVVGLI
MSMEG_5721|M.smegmatis_MC2_155 --------------MSEEAFIYEAIRTPRGKQRGGALNEIKPVNLVVGLI
Mvan_5067|M.vanbaalenii_PYR-1 --------------MSEEAFIYEAIRTPRGKQRNGALNEIKPVNLVVGLI
MAB_0850|M.abscessus_ATCC_1997 ---------------MTEAFIYEAVRTPRGKQRGGALNEIKPLNLVVGLI
TH_0255|M.thermoresistible__bu ---------------MRETVIVEAVRTAVGK-RNGALADVHAADLSAVVL
*:.* **:**. ** : *:* :::. .* . ::
MUL_0296|M.ulcerans_Agy99 EELRKRNPDLDENLISDVILGCVSPVGDQGGDIARAAVLASGMPVTSGGV
MMAR_4677|M.marinum_M EELRKRNPDLDENLISDVILGCVSPVGDQGGDIARAAVLASGMPVTSGGV
Mb0882|M.bovis_AF2122/97 DELRKRHPDLDENLISDVILGCVSPVGDQGGDIARAAVLASGMPVTSGGV
Rv0859|M.tuberculosis_H37Rv DELRKRHPDLDENLISDVILGCVSPVGDQGGDIARAAVLASGMPVTSGGV
MLBr_02162|M.leprae_Br4923 EELRKRNPDLDENLISDVILGCVSPVGDQGGDIARATVLAAGLPVTAGGV
MAV_0980|M.avium_104 DELRRRYPDLDETLISDMILGVVSPVGDQGGDIARTAVLAAGLPETTGGV
Mflv_1691|M.gilvum_PYR-GCK DEMRTRFPDLDENLISDLILGVVSPVGDQGGDIARTAGLVAKLPETTGGV
MSMEG_5721|M.smegmatis_MC2_155 DELRVRFPNLDETLISDLVLGVVSPVGDQGGDIARTATLVANLPETTGGV
Mvan_5067|M.vanbaalenii_PYR-1 DEMRVRFPDLDENLISDLILGCVSPVGDQGGDIARTAGLVAGLPDTTGGF
MAB_0850|M.abscessus_ATCC_1997 EEIRHRHPDLDENLISDVVLGVVSPVGDQGGDIARTAVLAAGLPDTTGGV
TH_0255|M.thermoresistible__bu SELVER-AGIDPELIDDVIWGCVSQVGDQSSNIGRYAVLAAGWPETIPGT
.*: * ..:* **.*:: * ** ****..:*.* : *.: * * *
MUL_0296|M.ulcerans_Agy99 QLNRFCASGLEAVNMAAQKVRSGWDDLVLAGGVESMSRVPMGAD---GGA
MMAR_4677|M.marinum_M QLNRFCASGLEAVNIAAQKVRSGWDDLVLAGGVESMSRVPMGAD---GGA
Mb0882|M.bovis_AF2122/97 QLNRFCASGLEAVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
Rv0859|M.tuberculosis_H37Rv QLNRFCASGLEAVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
MLBr_02162|M.leprae_Br4923 QLNRFCASGLEAVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
MAV_0980|M.avium_104 QLNRFCASGLEAVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
Mflv_1691|M.gilvum_PYR-GCK QLNRFCASGLEAVNMAAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
MSMEG_5721|M.smegmatis_MC2_155 QLNRFCASGLEAVNIAAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
Mvan_5067|M.vanbaalenii_PYR-1 QLNRFCASGLEAVNLGAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
MAB_0850|M.abscessus_ATCC_1997 QLNRFCASGLEAINIGAQKVRSGWDDLVLAGGVESMSRVPMGSD---GGA
TH_0255|M.thermoresistible__bu TVNRACGSSQQALDFAVHAVMSGQQDVVVAGGVEVMSRVPLGAARATGMP
:** *.*. :*:: ..: * ** :*:*:***** *****:*: * .
MUL_0296|M.ulcerans_Agy99 MGLDPVTNYDVMFVPQGIGADLIATIEGFSRDDVDAYALRSQEKAAEAWS
MMAR_4677|M.marinum_M MGLDPVTNYDVMFVPQGIGADLIATIEGFSRDDVDAYALRSQEKAAEAWS
Mb0882|M.bovis_AF2122/97 MGLDPATNYDVMFVPQGIGADLIATIEGFSREDVDAYALRSQQKAAEAWS
Rv0859|M.tuberculosis_H37Rv MGLDPATNYDVMFVPQSIGADLIATIEGFSREDVDAYALRSQQKAAEAWS
MLBr_02162|M.leprae_Br4923 MGLDPATNYDVMFVPQGIGADLIATIEGFSRDDVDVYALRSQEKAAAAWS
MAV_0980|M.avium_104 WATDPETNYRIGFVPQGIGADLIATLEGFSREDVDAYALRSQQKAAAAWS
Mflv_1691|M.gilvum_PYR-GCK WAADPETNYRIGFVPQGIGADLIATIEGFSREDVDAYAARSQERAAAAWS
MSMEG_5721|M.smegmatis_MC2_155 WAGDPETNYRIGFVPQGIGADLIATIEGFSREDVDAYAARSQERAAAAWS
Mvan_5067|M.vanbaalenii_PYR-1 WAGDPETNYRIGFVPQGIGADLIATIEGFSREDVDAYAARSQERAAAAWA
MAB_0850|M.abscessus_ATCC_1997 WASDPATNYDVSFVPQGIGADLIATIEGFTRDDVDAYAARSQERAAAAWS
TH_0255|M.thermoresistible__bu YGPKVLERYDNFGFNQGISAELIAERWNLSRTRLDEYSAQSHERAAAAQS
. . .* . *.*.*:*** .::* :* *: :*:::** * :
MUL_0296|M.ulcerans_Agy99 GGYFAKSVVPVRDQNGLLILDHDEHMRPETTKEGLGKLKSAFEGLAAMGG
MMAR_4677|M.marinum_M GGYFAKSVVPVRDQNGLLILDHDEHMRPETTKEGLGKLKSAFEGLAAMGG
Mb0882|M.bovis_AF2122/97 GGYFAKSVVPVRDQNGLLILDHDEHMRPDTTKEGLAKLKPAFEGLAALGG
Rv0859|M.tuberculosis_H37Rv GGYFAKSVVPVRDQNGLLILDHDEHMRPDTTKEGLAKLKPAFEGLAALGG
MLBr_02162|M.leprae_Br4923 GGYFAKSVVPVRDQNGLLILDHDEHMRPDTSKEGLAKLKPAFEGLAAMGG
MAV_0980|M.avium_104 GGYFAKSVVPVRDQNGLVILDHDEHMRPDTTMEGLAKLKTAFDGVGEMGG
Mflv_1691|M.gilvum_PYR-GCK GGYFAKSVVPVKDQNGLVVLDHDEHIRPGTTIESLGKLKSAFEGVGAMGG
MSMEG_5721|M.smegmatis_MC2_155 GGYFSKSVVPVKDQNGLVVLDHDEHMRPGTTVESLGKLKPAFEGVGAMGG
Mvan_5067|M.vanbaalenii_PYR-1 GGYFAKSVVPVKDQNGLVVLDHDEHMRPGSTVESLGKLKTAFDGIGAMGG
MAB_0850|M.abscessus_ATCC_1997 GGYFAKSVVPVKDQNGLLVLDHDEHMRPGTTAADLGKLKPAFEGLAALGG
TH_0255|M.thermoresistible__bu SGAFKDQIVPVFTDD--AVVTEDEGIRRGTTVEKLATLKPAFT-------
.* * ..:*** :: :: .** :* :: *..**.**
MUL_0296|M.ulcerans_Agy99 FDDVALQKYHWVEKINHVHTGGNSSGIVDGAAIVMVGSEKAGAAINATPR
MMAR_4677|M.marinum_M FDDVALQKYHWVEKINHVHTGGNSSGIVDGAAIVMVGSEKAGAAINATPR
Mb0882|M.bovis_AF2122/97 FDDVALQKYHWVEKINHVHTGGNSSGIVDGAALVMIGSAAAGKLQGLTPR
Rv0859|M.tuberculosis_H37Rv FDDVALQKYHWVEKINHVHTGGNSSGIVDGAALVMIGSAAAGKLQGLTPR
MLBr_02162|M.leprae_Br4923 FDDVALQKYHWVEKINHVHTGGNSSGIVDGAALVMIGSEAVGKSQGLTPR
MAV_0980|M.avium_104 FDDVALQKYHWVEKINHVHTGGNSSGIVDGAALVLVGSEKAGKSQGLTPR
Mflv_1691|M.gilvum_PYR-GCK FDDVALQKYHFVEKINHVHTGGNSSGIVDGAALVLIGSESAGTSQGLTPR
MSMEG_5721|M.smegmatis_MC2_155 FDDVALQKYHSIEKINHVHTGGNSSGIVDGAALVLIGSESAGKAAGLTPR
Mvan_5067|M.vanbaalenii_PYR-1 FDDVALQKYHFVEKINHVHTGGNSSGIVDGAALLLIGSEAAGKSQGLTPR
MAB_0850|M.abscessus_ATCC_1997 FDDVARIKYHYVEKINHVHTGGNSSGIVDGAALVLIGSEEAGKSQGLTPR
TH_0255|M.thermoresistible__bu --------------ENGVIHAGNSSQISDGAAALLVMTAETAVTLGLTPI
* * .**** * **** ::: : .. . **
MUL_0296|M.ulcerans_Agy99 ARIVATATSGADPVIMLTGPTPAARKVLDRAGLTVDDIDLFELNEAFASV
MMAR_4677|M.marinum_M ARIVATATSGADPVIMLTGPTPAARKVLDRAGLTVDDIDLFELNEAFASV
Mb0882|M.bovis_AF2122/97 ARIVATATSGADPVIMLTGPTPATRKVLDRAGLTVDDIDLFELNEAFASV
Rv0859|M.tuberculosis_H37Rv ARIVATATSGADPVIMLTGPTPATRKVLDRAGLTVDDIDLFELNEAFASV
MLBr_02162|M.leprae_Br4923 ARMVATATSGADPVIMLTGPTPATRKVLDRAGLTVDDIDLFELNEAFASV
MAV_0980|M.avium_104 ARIVATATSGSDPVIMLTGPTPATRKVLDRAGLTIDDIDLFELNEAFASV
Mflv_1691|M.gilvum_PYR-GCK ARIVATATSGADPVIMLTGPTPATKKVLDRAGLTVDDIDLFELNEAFASV
MSMEG_5721|M.smegmatis_MC2_155 ARVVATATSGADPVIMLTGPTPATRKVLDRAGLTVDDIDLFELNEAFASV
Mvan_5067|M.vanbaalenii_PYR-1 ARIVATATSGADPVIMLTGPTPATQKVLDRAGLTVDDIDLFELNEAFASV
MAB_0850|M.abscessus_ATCC_1997 ARIVATATSGSEPLIMLTGPTPATKKVLDRAGLTVDDIDLFELNEAFASV
TH_0255|M.thermoresistible__bu VRYRAGAVTGADPVLMLTGPIPATEKVLRKTGLNIDDIGVFEVNEAFAPV
.* * *.:*::*::***** **:.*** ::**.:***.:**:*****.*
MUL_0296|M.ulcerans_Agy99 VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMILGTMVDELERRNARR
MMAR_4677|M.marinum_M VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMILGTMVDELERRNARR
Mb0882|M.bovis_AF2122/97 VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMILGTMVDELERRNARR
Rv0859|M.tuberculosis_H37Rv VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMILGTMVDELERRNARR
MLBr_02162|M.leprae_Br4923 VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMILGTMVDELERCNARR
MAV_0980|M.avium_104 VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMITGTMVDELERRNARR
Mflv_1691|M.gilvum_PYR-GCK VLKFQKDLGIPDEKLNVNGGAIAMGHPLGATGAMITGTMVDELERRGARR
MSMEG_5721|M.smegmatis_MC2_155 VLKFQKDLGIPDEKLNVNGGAIAMGHPLGATGAMITGTMVDELERRGARR
Mvan_5067|M.vanbaalenii_PYR-1 VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMITGTMVDELERRGAKR
MAB_0850|M.abscessus_ATCC_1997 VLKFQKDLNIPDEKLNVNGGAIAMGHPLGATGAMITGTMVDELERRGLKR
TH_0255|M.thermoresistible__bu PLAWQAETGADPAKLNPLGGAIALGHPLGASGAVLMTRMVHHMRANGIRY
* :* : . *** *****:******:**:: **..:. . :
MUL_0296|M.ulcerans_Agy99 ALVTLCIGGGMGAATIIERV-
MMAR_4677|M.marinum_M ALVTLCIGGGMGVATIIERV-
Mb0882|M.bovis_AF2122/97 ALITLCIGGGMGVATIIERV-
Rv0859|M.tuberculosis_H37Rv ALITLCIGGGMGVATIIERV-
MLBr_02162|M.leprae_Br4923 ALVTLCIGAGMGVATIIERV-
MAV_0980|M.avium_104 ALITLCIGGGMGVATIIERV-
Mflv_1691|M.gilvum_PYR-GCK ALITLCVGGGMGVATIIERV-
MSMEG_5721|M.smegmatis_MC2_155 ALITLCVGGGMGVATIIERV-
Mvan_5067|M.vanbaalenii_PYR-1 ALMTLCVGGGMGVATIIERV-
MAB_0850|M.abscessus_ATCC_1997 ALITLCVGGGMGVATIIERV-
TH_0255|M.thermoresistible__bu GLQTMCEGGGTANATVVELLA
.* *:* *.* . **::* :